DISEASE EPIDEMIOLOGY
Population-level descriptive epidemiology
These analyses include an assessment of the incidence or prevalence of specific conditions, eventually stratified by time or by population subgroups like age or sex.
A typical population-level descriptive epidemiology study will have an objective like:
To estimate the monthly incidence rate and point prevalence of disease A stratified by sex and age.
Report Contents
The report will include an executive summary and the following tables and figures:
- Table 1: Number of participants and total number of incident and/or prevalent cases in each data source during the study period. Number of participants per pre-specified strata will be included where necessary/applicable.
CohortCharacteristics::tableCharacteristics(
result,
type = "gt",
header = c("cdm_name", "cohort_name"),
groupColumn = character(),
hide = c(additionalColumns(result), settingsColumns(result)),
.options = list(style = "darwin")
)|
CDM name
|
|||||
|---|---|---|---|---|---|
|
PP_MOCK
|
|||||
| Variable name | Variable level | Estimate name |
Cohort name
|
||
| cohort_1 | cohort_2 | cohort_3 | |||
| Number records | - | N | 6 | 2 | 2 |
| Number subjects | - | N | 6 | 2 | 2 |
| Cohort start date | - | Median [Q25 - Q75] | 1976-02-17 [1926-12-14 - 1983-07-28] | 1955-12-29 [1943-09-07 - 1968-04-20] | 1991-04-01 [1980-01-29 - 2002-06-01] |
| Range | 1907-01-04 to 1991-11-21 | 1931-05-17 to 1980-08-11 | 1968-11-27 to 2013-08-02 | ||
| Cohort end date | - | Median [Q25 - Q75] | 1983-05-14 [1930-11-19 - 1990-08-18] | 1958-08-28 [1945-02-08 - 1972-03-17] | 1995-05-06 [1986-03-13 - 2004-06-27] |
| Range | 1912-07-22 to 1997-10-31 | 1931-07-22 to 1985-10-05 | 1977-01-19 to 2013-08-19 | ||
| Age | - | Median [Q25 - Q75] | 7 [3 - 18] | 12 [10 - 15] | 24 [19 - 30] |
| Mean (SD) | 12.17 (12.59) | 12.50 (7.78) | 24.50 (16.26) | ||
| Range | 2 to 33 | 7 to 18 | 13 to 36 | ||
| Sex | Female | N (%) | 4 (66.67%) | 2 (100.00%) | 1 (50.00%) |
| Male | N (%) | 2 (33.33%) | - | 1 (50.00%) | |
| Prior observation | - | Median [Q25 - Q75] | 2,850 [1,354 - 7,065] | 4,745 [3,719 - 5,771] | 9,220 [7,150 - 11,291] |
| Mean (SD) | 4,672.67 (4,637.15) | 4,745.00 (2,901.97) | 9,220.50 (5,856.97) | ||
| Range | 733 to 12,276 | 2,693 to 6,797 | 5,079 to 13,362 | ||
| Future observation | - | Median [Q25 - Q75] | 7,254 [4,476 - 12,970] | 2,052 [1,789 - 2,314] | 3,466 [2,248 - 4,685] |
| Mean (SD) | 8,389.17 (5,498.88) | 2,051.50 (743.17) | 3,466.50 (3,447.15) | ||
| Range | 2,246 to 15,156 | 1,526 to 2,577 | 1,029 to 5,904 | ||
| Days in cohort | - | Median [Q25 - Q75] | 2,366 [2,063 - 2,578] | 974 [521 - 1,428] | 1,497 [758 - 2,236] |
| Mean (SD) | 2,179.50 (633.44) | 974.50 (1,283.40) | 1,497.00 (2,091.62) | ||
| Range | 1,007 to 2,727 | 67 to 1,882 | 18 to 2,976 | ||
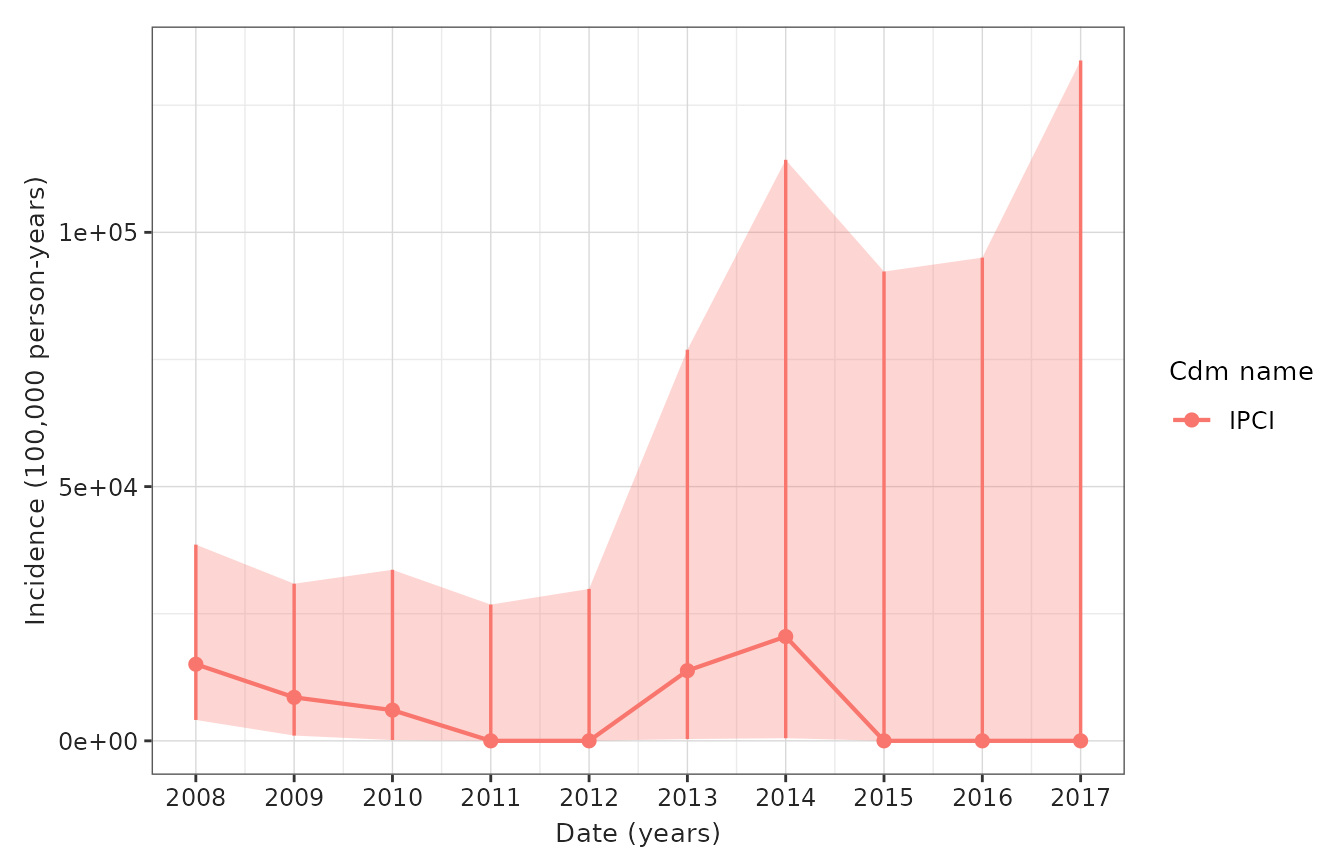
- Figure 1: Incidence rate/s of disease over calendar time (per month/year) overall
IncidencePrevalence::plotIncidence(
result = incidence_result_figure_1,
x = "incidence_start_date",
y = "incidence_100000_pys",
line = TRUE,
point = TRUE,
ribbon = TRUE,
ymin = "incidence_100000_pys_95CI_lower",
ymax = "incidence_100000_pys_95CI_upper",
facet = NULL,
colour = "cdm_name"
)
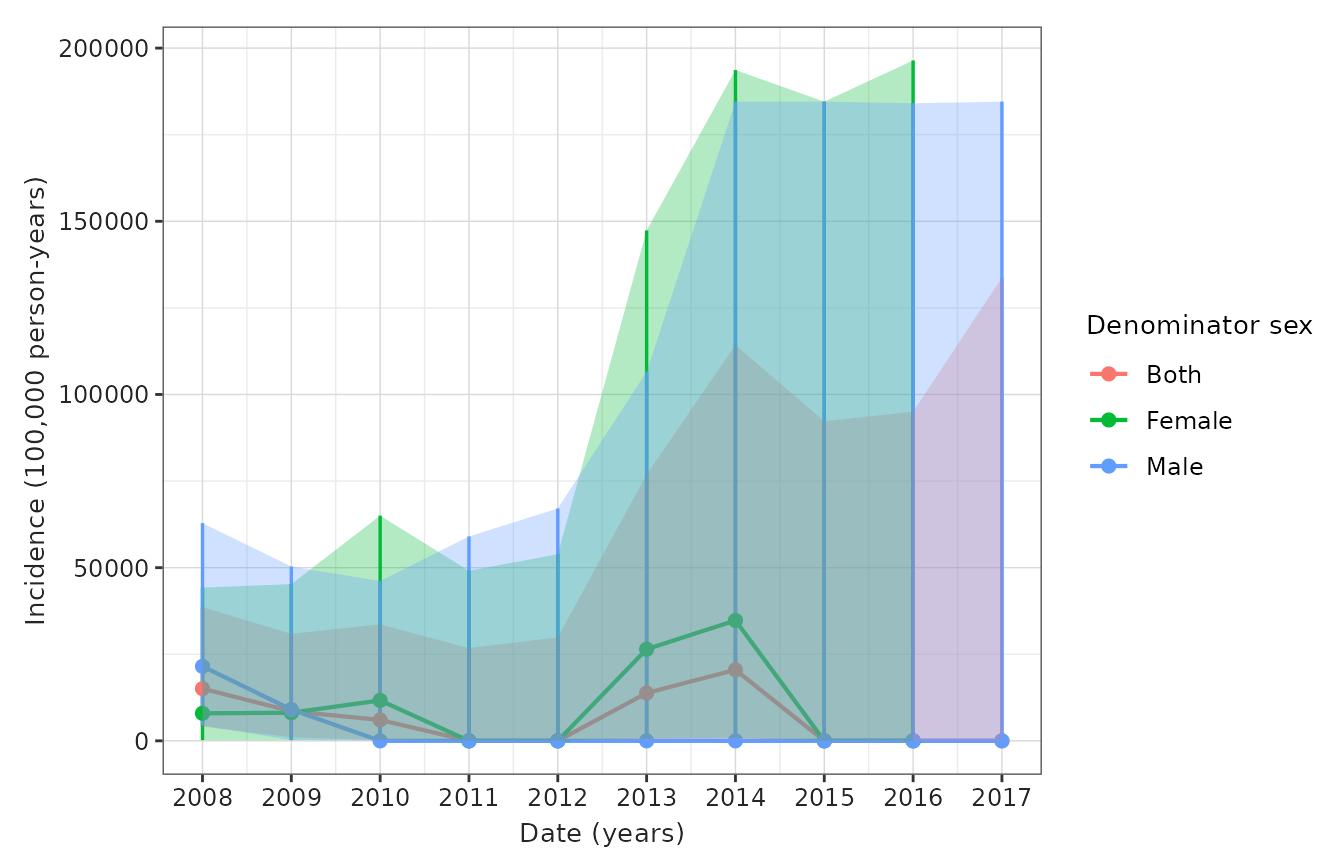
- Figure 2: Incidence rate/s of disease over calendar time (per month/year) stratified by sex and age (or other pre-specified criterion/a)
IncidencePrevalence::plotIncidence(
result = incidence_result_figure_2a,
x = "incidence_start_date",
y = "incidence_100000_pys",
line = TRUE,
point = TRUE,
ribbon = TRUE,
ymin = "incidence_100000_pys_95CI_lower",
ymax = "incidence_100000_pys_95CI_upper",
facet = NULL,
colour = "denominator_sex"
)
IncidencePrevalence::plotIncidence(
result = incidence_result_figure_2b,
x = "incidence_start_date",
y = "incidence_100000_pys",
line = TRUE,
point = TRUE,
ribbon = TRUE,
ymin = "incidence_100000_pys_95CI_lower",
ymax = "incidence_100000_pys_95CI_upper",
facet = NULL,
colour = "denominator_age_group"
)
- Table 2: Numbers reported in figures 1 and 2
IncidencePrevalence::tableIncidence(
result = incidence_result_table_2,
type = "gt",
header = c("estimate_name"),
groupColumn = c("cdm_name", "outcome_cohort_name"),
settingsColumn = c("denominator_age_group", "denominator_sex"),
hide = c("denominator_cohort_name", "analysis_interval"),
.options = list(style = "darwin")
)| Incidence start date | Incidence end date | Denominator age group | Denominator sex |
Estimate name
|
|||
|---|---|---|---|---|---|---|---|
| Denominator (N) | Person-years | Outcome (N) | Incidence 100,000 person-years [95% CI] | ||||
| IPCI; cohort_1 | |||||||
| 2008-01-01 | 2008-12-31 | 0 to 64 | Both | 32 | 26.54 | 4 | 15,069.89 (4,106.04 - 38,584.89) |
| 2009-01-01 | 2009-12-31 | 0 to 64 | Both | 30 | 23.38 | 2 | 8,552.86 (1,035.79 - 30,895.86) |
| 2010-01-01 | 2010-12-31 | 0 to 64 | Both | 22 | 16.56 | 1 | 6,037.19 (152.85 - 33,637.07) |
| 2011-01-01 | 2011-12-31 | 0 to 64 | Both | 16 | 13.77 | 0 | 0.00 (0.00 - 26,797.03) |
| 2012-01-01 | 2012-12-31 | 0 to 64 | Both | 13 | 12.35 | 0 | 0.00 (0.00 - 29,874.31) |
| 2013-01-01 | 2013-12-31 | 0 to 64 | Both | 11 | 7.24 | 1 | 13,804.53 (349.50 - 76,913.91) |
| 2014-01-01 | 2014-12-31 | 0 to 64 | Both | 5 | 4.88 | 1 | 20,508.61 (519.23 - 114,266.68) |
| 2015-01-01 | 2015-12-31 | 0 to 64 | Both | 4 | 4.00 | 0 | 0.00 (0.00 - 92,291.20) |
| 2016-01-01 | 2016-12-31 | 0 to 64 | Both | 5 | 3.88 | 0 | 0.00 (0.00 - 95,025.23) |
| 2017-01-01 | 2017-12-31 | 0 to 64 | Both | 3 | 2.76 | 0 | 0.00 (0.00 - 133,800.49) |
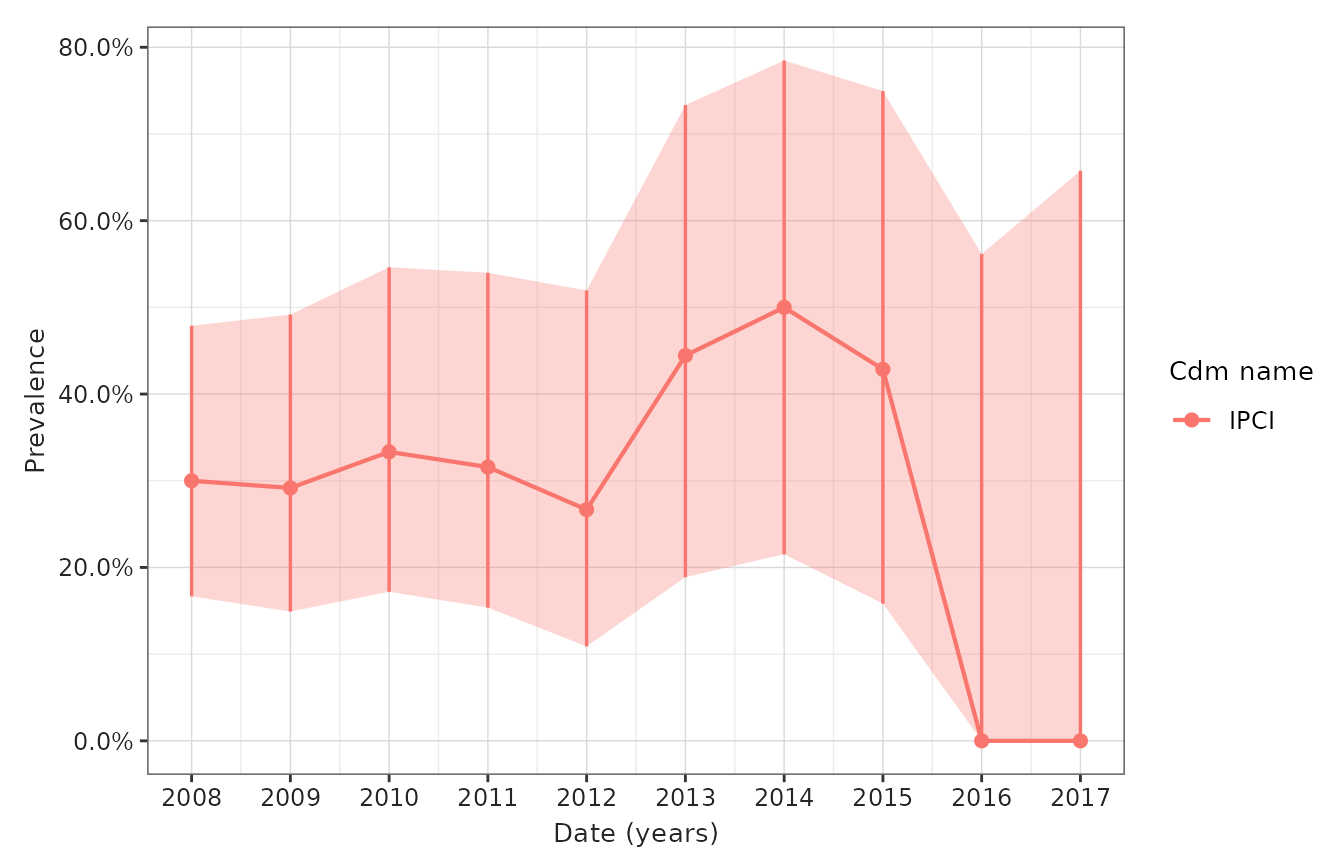
- Figure 3: Prevalence of disease over calendar time (per month/year) overall
IncidencePrevalence::plotPrevalence(
result = prevalence_result_figure_1,
x = "prevalence_start_date",
y = "prevalence",
line = TRUE,
point = TRUE,
ribbon = TRUE,
ymin = "prevalence_95CI_lower",
ymax = "prevalence_95CI_upper",
facet = NULL,
colour = "cdm_name"
)
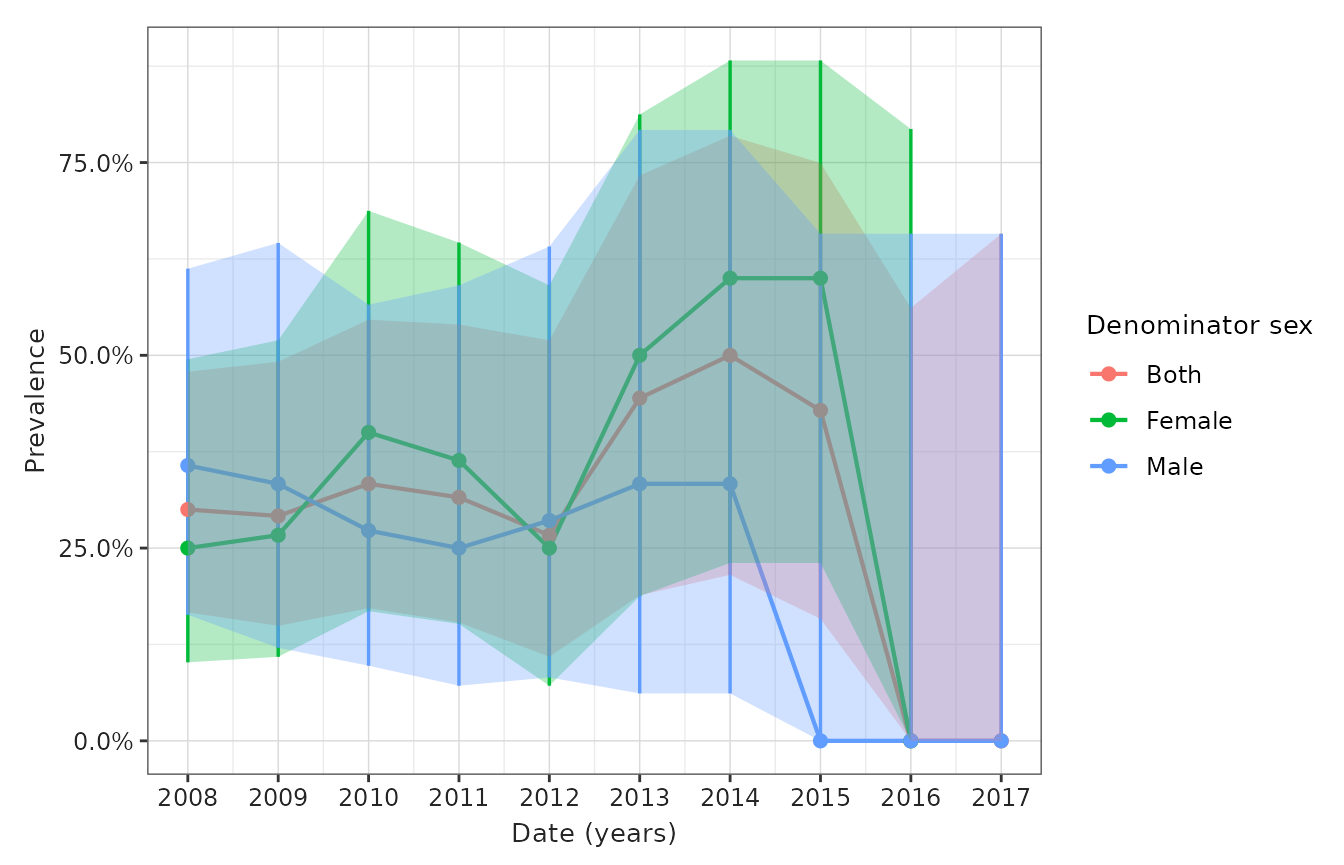
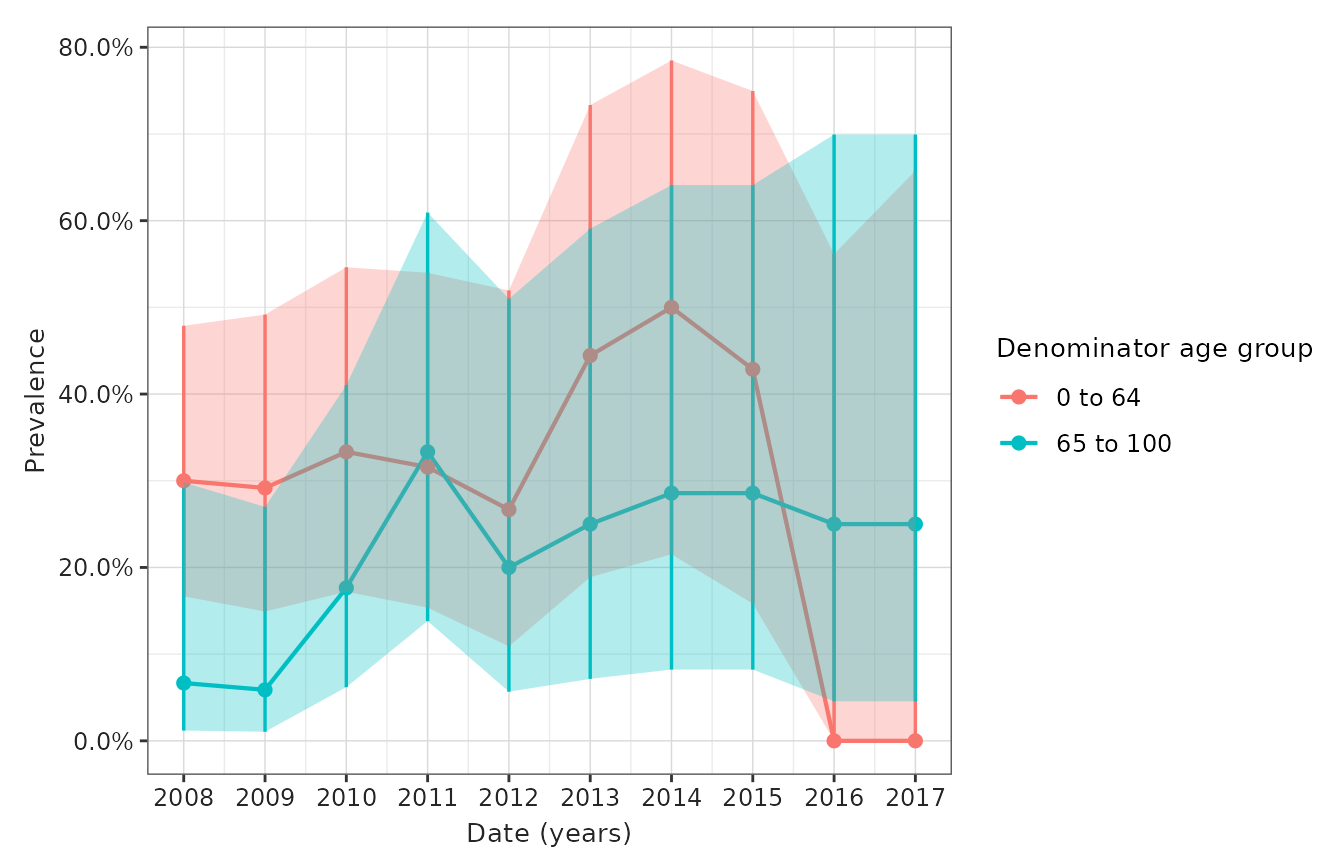
- Figure 4: Prevalence of disease over calendar time (per month/year) stratified by sex and age (or other pre-specified criterion/a)
IncidencePrevalence::plotPrevalence(
result = prevalence_result_figure_2a,
x = "prevalence_start_date",
y = "prevalence",
line = TRUE,
point = TRUE,
ribbon = TRUE,
ymin = "prevalence_95CI_lower",
ymax = "prevalence_95CI_upper",
facet = NULL,
colour = "denominator_sex"
)
IncidencePrevalence::plotPrevalence(
result = prevalence_result_figure_2b,
x = "prevalence_start_date",
y = "prevalence",
line = TRUE,
point = TRUE,
ribbon = TRUE,
ymin = "prevalence_95CI_lower",
ymax = "prevalence_95CI_upper",
facet = NULL,
colour = "denominator_age_group"
)
- Table 3: Numbers reported in Figures 3 and 4.
IncidencePrevalence::tablePrevalence(
result = prevalence_result_table_2,
type = "gt",
header = c("estimate_name"),
groupColumn = c("cdm_name", "outcome_cohort_name"),
settingsColumn = c("denominator_age_group", "denominator_sex"),
hide = c("denominator_cohort_name", "analysis_interval"),
.options = list(style = "darwin")
)| Prevalence start date | Prevalence end date | Denominator age group | Denominator sex |
Estimate name
|
||
|---|---|---|---|---|---|---|
| Denominator (N) | Outcome (N) | Prevalence [95% CI] | ||||
| IPCI; cohort_1 | ||||||
| 2008-01-01 | 2008-12-31 | 0 to 64 | Both | 30 | 9 | 0.30 (0.17 - 0.48) |
| 2009-01-01 | 2009-12-31 | 0 to 64 | Both | 24 | 7 | 0.29 (0.15 - 0.49) |
| 2010-01-01 | 2010-12-31 | 0 to 64 | Both | 21 | 7 | 0.33 (0.17 - 0.55) |
| 2011-01-01 | 2011-12-31 | 0 to 64 | Both | 19 | 6 | 0.32 (0.15 - 0.54) |
| 2012-01-01 | 2012-12-31 | 0 to 64 | Both | 15 | 4 | 0.27 (0.11 - 0.52) |
| 2013-01-01 | 2013-12-31 | 0 to 64 | Both | 9 | 4 | 0.44 (0.19 - 0.73) |
| 2014-01-01 | 2014-12-31 | 0 to 64 | Both | 8 | 4 | 0.50 (0.22 - 0.78) |
| 2015-01-01 | 2015-12-31 | 0 to 64 | Both | 7 | 3 | 0.43 (0.16 - 0.75) |
| 2016-01-01 | 2016-12-31 | 0 to 64 | Both | 3 | 0 | 0.00 (0.00 - 0.56) |
| 2017-01-01 | 2017-12-31 | 0 to 64 | Both | 2 | 0 | 0.00 (0.00 - 0.66) |
Patient-level characterization
These analyses include a descriptive analysis of a cohort of patients newly diagnosed with a specific condition of interest. This characterization typically includes pre-specified features (e.g. history of a certain list of diseases) as well as a list of the most commonly recorded codes in the patient records, and can be stratified by age, sex, or other characteristics.
A typical patient-level characterization study will have an objective that reads something like:
To characterise patients newly diagnosed with condition A, including an assessment of age, sex, and previous comorbidities, as well as a descriptive analysis of how they are treated in the year after diagnosis
Report Contents
The report will include an executive summary and the following tables and figures:
- Table 1. Pre-index characteristics of patients newly diagnosed with a condition of interest, at the time of diagnosis/recording. Number of participants per pre-specified strata will be included where necessary/applicable.
CohortCharacteristics::tableCharacteristics(
result = summarise_characteristics,
type = "gt",
header = c("cdm_name", "cohort_name"),
groupColumn = "variable_name",
hide = c("table", "window", "value", "table_name"),
.options = list(style = "darwin")
)|
CDM name
|
|||||
|---|---|---|---|---|---|
|
IPCI
|
|||||
| Variable level | Estimate name |
Cohort name
|
|||
| ankle_sprain | ankle_fracture | forearm_fracture | hip_fracture | ||
| Number records | |||||
| - | N | 1,915 | 464 | 569 | 138 |
| Number subjects | |||||
| - | N | 1,357 | 427 | 510 | 132 |
| Cohort start date | |||||
| - | Median [Q25 - Q75] | 1982-11-09 [1968-06-15 - 1999-04-13] | 1981-01-15 [1965-03-11 - 1997-08-03] | 1981-07-24 [1967-03-05 - 2000-12-16] | 1996-09-17 [1977-09-20 - 2010-06-22] |
| Range | 1912-02-25 to 2019-05-30 | 1911-09-07 to 2019-06-23 | 1917-08-16 to 2019-06-26 | 1927-12-14 to 2019-05-08 | |
| Cohort end date | |||||
| - | Median [Q25 - Q75] | 1982-12-10 [1968-07-06 - 1999-05-09] | 1981-02-28 [1965-04-11 - 1997-10-12] | 1981-08-23 [1967-04-10 - 2001-02-27] | 1996-11-16 [1977-12-04 - 2010-07-22] |
| Range | 1912-03-10 to 2019-05-30 | 1911-12-06 to 2019-06-24 | 1917-11-14 to 2019-06-26 | 1928-03-13 to 2019-06-07 | |
| Age | |||||
| - | Median [Q25 - Q75] | 21 [9 - 41] | 16 [9 - 43] | 17 [9 - 46] | 40 [13 - 66] |
| Mean (SD) | 26.63 (21.03) | 27.38 (24.70) | 28.69 (25.97) | 40.06 (28.82) | |
| Range | 0 to 105 | 0 to 107 | 0 to 106 | 1 to 108 | |
| Sex | |||||
| Female | N (%) | 954 (49.82%) | 238 (51.29%) | 286 (50.26%) | 74 (53.62%) |
| Male | N (%) | 961 (50.18%) | 226 (48.71%) | 283 (49.74%) | 64 (46.38%) |
| Prior observation | |||||
| - | Median [Q25 - Q75] | 7,833 [3,628 - 15,147] | 6,030 [3,360 - 16,032] | 6,289 [3,390 - 16,847] | 14,522 [4,801 - 24,401] |
| Mean (SD) | 9,918.17 (7,672.74) | 10,196.57 (9,011.31) | 10,670.43 (9,480.30) | 14,821.73 (10,521.89) | |
| Range | 299 to 38,429 | 299 to 39,430 | 299 to 38,943 | 390 to 39,792 | |
| Future observation | |||||
| - | Median [Q25 - Q75] | 12,868 [6,860 - 18,078] | 13,748 [6,878 - 19,331] | 13,165 [5,988 - 18,548] | 7,798 [2,874 - 14,913] |
| Mean (SD) | 12,865.11 (7,543.50) | 13,470.92 (8,215.96) | 12,913.27 (7,929.17) | 9,167.33 (7,160.81) | |
| Range | 0 to 38,403 | 1 to 39,051 | 0 to 36,654 | 0 to 29,045 | |
| Days in cohort | |||||
| - | Median [Q25 - Q75] | 22 [15 - 29] | 61 [31 - 91] | 61 [31 - 91] | 61 [31 - 91] |
| Mean (SD) | 25.02 (8.00) | 61.65 (25.38) | 62.16 (25.32) | 59.26 (24.79) | |
| Range | 1 to 37 | 2 to 92 | 1 to 91 | 1 to 91 | |
| Medications prior to index date | |||||
| Acetaminophen | N (%) | 1,530 (79.90%) | 357 (76.94%) | 447 (78.56%) | 119 (86.23%) |
| Warfarin | N (%) | 12 (0.63%) | 8 (1.72%) | 11 (1.93%) | 4 (2.90%) |
| Morphine | N (%) | 15 (0.78%) | 1 (0.22%) | 2 (0.35%) | 2 (1.45%) |
| Medications on index date | |||||
| Acetaminophen | N (%) | 773 (40.37%) | 240 (51.72%) | 264 (46.40%) | 90 (65.22%) |
| Morphine | N (%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| Warfarin | N (%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
- Table 2. Number and percentage with an outcome of interest in the x years following diagnosis.
CohortCharacteristics::tableTopLargeScaleCharacteristics(
result = lsc_characteristics,
topConcepts = 10,
type = "gt"
)|
Window
|
|||||
|---|---|---|---|---|---|
|
-inf to -1
|
0 to 0
|
||||
|
Table name
|
|||||
|
condition_occurrence
|
drug_exposure
|
procedure_occurrence
|
condition_occurrence
|
drug_exposure
|
|
| Top |
Type
|
||||
| event | episode | event | event | episode | |
| 1 | Viral sinusitis (40481087) 981 (72.3%) |
poliovirus vaccine, inactivated (40213160) 994 (73.2%) |
Suture open wound (4125906) 363 (26.8%) |
Sprain of ankle (81151) 1357 (100.0%) |
Aspirin 81 MG Oral Tablet (19059056) 470 (34.6%) |
| 2 | Otitis media (372328) 909 (67.0%) |
Aspirin 81 MG Oral Tablet (19059056) 842 (62.0%) |
Bone immobilization (4170947) 356 (26.2%) |
Acetaminophen 325 MG Oral Tablet (1127433) 330 (24.3%) |
|
| 3 | Acute viral pharyngitis (4112343) 845 (62.3%) |
Acetaminophen 325 MG Oral Tablet (1127433) 737 (54.3%) |
Sputum examination (4151422) 282 (20.8%) |
Acetaminophen 160 MG Oral Tablet (1127078) 199 (14.7%) |
|
| 4 | Acute bronchitis (260139) 767 (56.5%) |
Acetaminophen 160 MG Oral Tablet (1127078) 559 (41.2%) |
Plain chest X-ray (4163872) 137 (10.1%) |
Ibuprofen 200 MG Oral Tablet (19078461) 192 (14.2%) |
|
| 5 | Streptococcal sore throat (28060) 499 (36.8%) |
Amoxicillin 250 MG / Clavulanate 125 MG Oral Tablet (1713671) 499 (36.8%) |
|||
| 6 | Osteoarthritis (80180) 283 (20.9%) |
Penicillin V Potassium 250 MG Oral Tablet (19133873) 491 (36.2%) |
|||
| 7 | Concussion with no loss of consciousness (378001) 185 (13.6%) |
Penicillin G 375 MG/ML Injectable Solution (19006318) 384 (28.3%) |
|||
| 8 | Acute bacterial sinusitis (4294548) 168 (12.4%) |
Acetaminophen 21.7 MG/ML / Dextromethorphan Hydrobromide 1 MG/ML / doxylamine succinate 0.417 MG/ML Oral Solution (40229134) 296 (21.8%) |
|||
| 9 | Sinusitis (4283893) 166 (12.2%) |
tetanus and diphtheria toxoids, adsorbed, preservative free, for adult use (40213227) 288 (21.2%) |
|||
| 10 | Chronic sinusitis (257012) 162 (11.9%) |
hepatitis B vaccine, adult dosage (40213306) 226 (16.6%) |
|||
- Figure 1. Kaplan-Meier or Cumulative Incidence Function plots of the probability of a pre-specified outcome following index diagnosis of the condition of interest.
CohortSurvival::tableSurvival(
x = single_event_data,
times = NULL,
timeScale = "days",
header = c("estimate"),
type = "gt",
groupColumn = NULL,
.options = list(style = "darwin")
)
CohortSurvival::riskTable(
x = competing_risk_data,
eventGap = NULL,
header = c("estimate"),
type = "gt",
groupColumn = NULL,
.options = list(style = "darwin")
)| CDM name | Target cohort | Sex | Outcome type | Outcome name | Time | Event gap |
Estimate name
|
||
|---|---|---|---|---|---|---|---|---|---|
| Number at risk | Number events | Number censored | |||||||
| IPCI | mgus_diagnosis | overall | outcome | progression | 0 | 30 | 1,384 | 0 | 0 |
| 30 | 30 | 1,090 | 25 | 3 | |||||
| 60 | 30 | 874 | 19 | 27 | |||||
| 90 | 30 | 635 | 19 | 77 | |||||
| 120 | 30 | 424 | 13 | 74 | |||||
| 150 | 30 | 288 | 9 | 52 | |||||
| 180 | 30 | 178 | 10 | 52 | |||||
| 210 | 30 | 103 | 4 | 54 | |||||
| 240 | 30 | 57 | 2 | 32 | |||||
| 270 | 30 | 29 | 1 | 18 | |||||
| 300 | 30 | 16 | 1 | 10 | |||||
| 330 | 30 | 7 | 1 | 6 | |||||
| 360 | 30 | 3 | 1 | 3 | |||||
| 390 | 30 | 2 | 1 | 0 | |||||
| 420 | 30 | 1 | 0 | 1 | |||||
| 424 | 30 | 1 | 0 | 0 | |||||
| competing_outcome | death_cohort | 0 | 30 | 1,384 | 0 | 0 | |||
| 30 | 30 | 1,090 | 275 | 3 | |||||
| 60 | 30 | 874 | 170 | 27 | |||||
| 90 | 30 | 635 | 147 | 77 | |||||
| 120 | 30 | 424 | 113 | 74 | |||||
| 150 | 30 | 288 | 77 | 52 | |||||
| 180 | 30 | 178 | 47 | 52 | |||||
| 210 | 30 | 103 | 16 | 54 | |||||
| 240 | 30 | 57 | 12 | 32 | |||||
| 270 | 30 | 29 | 8 | 18 | |||||
| 300 | 30 | 16 | 1 | 10 | |||||
| 330 | 30 | 7 | 2 | 6 | |||||
| 360 | 30 | 3 | 0 | 3 | |||||
| 390 | 30 | 2 | 0 | 0 | |||||
| 420 | 30 | 1 | 0 | 1 | |||||
| 424 | 30 | 1 | 1 | 0 | |||||
| F | outcome | progression | 0 | 30 | 631 | 0 | 0 | ||
| 30 | 30 | 511 | 16 | 2 | |||||
| 60 | 30 | 431 | 8 | 10 | |||||
| 90 | 30 | 321 | 10 | 32 | |||||
| 120 | 30 | 214 | 7 | 44 | |||||
| 150 | 30 | 143 | 4 | 24 | |||||
| 180 | 30 | 88 | 4 | 27 | |||||
| 210 | 30 | 54 | 1 | 25 | |||||
| 240 | 30 | 33 | 1 | 16 | |||||
| 270 | 30 | 18 | 0 | 11 | |||||
| 300 | 30 | 11 | 1 | 5 | |||||
| 330 | 30 | 4 | 1 | 4 | |||||
| 360 | 30 | 2 | 1 | 1 | |||||
| 390 | 30 | 1 | 1 | 0 | |||||
| 394 | 30 | 1 | 0 | 1 | |||||
| M | outcome | progression | 0 | 30 | 753 | 0 | 0 | ||
| 30 | 30 | 579 | 9 | 1 | |||||
| 60 | 30 | 443 | 11 | 17 | |||||
| 90 | 30 | 314 | 9 | 45 | |||||
| 120 | 30 | 210 | 6 | 30 | |||||
| 150 | 30 | 145 | 5 | 28 | |||||
| 180 | 30 | 90 | 6 | 25 | |||||
| 210 | 30 | 49 | 3 | 29 | |||||
| 240 | 30 | 24 | 1 | 16 | |||||
| 270 | 30 | 11 | 1 | 7 | |||||
| 300 | 30 | 5 | 0 | 5 | |||||
| 330 | 30 | 3 | 0 | 2 | |||||
| 360 | 30 | 1 | 0 | 2 | |||||
| 390 | 30 | 1 | 0 | 0 | |||||
| 420 | 30 | 1 | 0 | 0 | |||||
| 424 | 30 | 1 | 0 | 0 | |||||
| F | competing_outcome | death_cohort | 0 | 30 | 631 | 0 | 0 | ||
| 30 | 30 | 511 | 107 | 2 | |||||
| 60 | 30 | 431 | 60 | 10 | |||||
| 90 | 30 | 321 | 67 | 32 | |||||
| 120 | 30 | 214 | 56 | 44 | |||||
| 150 | 30 | 143 | 42 | 24 | |||||
| 180 | 30 | 88 | 23 | 27 | |||||
| 210 | 30 | 54 | 8 | 25 | |||||
| 240 | 30 | 33 | 6 | 16 | |||||
| 270 | 30 | 18 | 3 | 11 | |||||
| 300 | 30 | 11 | 0 | 5 | |||||
| 330 | 30 | 4 | 2 | 4 | |||||
| 360 | 30 | 2 | 0 | 1 | |||||
| 390 | 30 | 1 | 0 | 0 | |||||
| 394 | 30 | 1 | 0 | 1 | |||||
| M | competing_outcome | death_cohort | 0 | 30 | 753 | 0 | 0 | ||
| 30 | 30 | 579 | 168 | 1 | |||||
| 60 | 30 | 443 | 110 | 17 | |||||
| 90 | 30 | 314 | 80 | 45 | |||||
| 120 | 30 | 210 | 57 | 30 | |||||
| 150 | 30 | 145 | 35 | 28 | |||||
| 180 | 30 | 90 | 24 | 25 | |||||
| 210 | 30 | 49 | 8 | 29 | |||||
| 240 | 30 | 24 | 6 | 16 | |||||
| 270 | 30 | 11 | 5 | 7 | |||||
| 300 | 30 | 5 | 1 | 5 | |||||
| 330 | 30 | 3 | 0 | 2 | |||||
| 360 | 30 | 1 | 0 | 2 | |||||
| 390 | 30 | 1 | 0 | 0 | |||||
| 420 | 30 | 1 | 0 | 0 | |||||
| 424 | 30 | 1 | 1 | 0 | |||||
- Table 3. Number and percentage treated with a pre-specified medicine or list of medicines within a pre-specified time period following an index diagnosis.
CohortCharacteristics::tableCharacteristics(
result = summarise_characteristics,
type = "gt",
header = c("cdm_name", "cohort_name"),
groupColumn = "variable_name",
hide = c("table", "window", "value", "table_name"),
.options = list(style = "darwin")
)|
CDM name
|
|||||
|---|---|---|---|---|---|
|
IPCI
|
|||||
| Variable level | Estimate name |
Cohort name
|
|||
| ankle_sprain | ankle_fracture | forearm_fracture | hip_fracture | ||
| Number records | |||||
| - | N | 1,915 | 464 | 569 | 138 |
| Number subjects | |||||
| - | N | 1,357 | 427 | 510 | 132 |
| Cohort start date | |||||
| - | Median [Q25 - Q75] | 1982-11-09 [1968-06-15 - 1999-04-13] | 1981-01-15 [1965-03-11 - 1997-08-03] | 1981-07-24 [1967-03-05 - 2000-12-16] | 1996-09-17 [1977-09-20 - 2010-06-22] |
| Range | 1912-02-25 to 2019-05-30 | 1911-09-07 to 2019-06-23 | 1917-08-16 to 2019-06-26 | 1927-12-14 to 2019-05-08 | |
| Cohort end date | |||||
| - | Median [Q25 - Q75] | 1982-12-10 [1968-07-06 - 1999-05-09] | 1981-02-28 [1965-04-11 - 1997-10-12] | 1981-08-23 [1967-04-10 - 2001-02-27] | 1996-11-16 [1977-12-04 - 2010-07-22] |
| Range | 1912-03-10 to 2019-05-30 | 1911-12-06 to 2019-06-24 | 1917-11-14 to 2019-06-26 | 1928-03-13 to 2019-06-07 | |
| Age | |||||
| - | Median [Q25 - Q75] | 21 [9 - 41] | 16 [9 - 43] | 17 [9 - 46] | 40 [13 - 66] |
| Mean (SD) | 26.63 (21.03) | 27.38 (24.70) | 28.69 (25.97) | 40.06 (28.82) | |
| Range | 0 to 105 | 0 to 107 | 0 to 106 | 1 to 108 | |
| Sex | |||||
| Female | N (%) | 954 (49.82%) | 238 (51.29%) | 286 (50.26%) | 74 (53.62%) |
| Male | N (%) | 961 (50.18%) | 226 (48.71%) | 283 (49.74%) | 64 (46.38%) |
| Prior observation | |||||
| - | Median [Q25 - Q75] | 7,833 [3,628 - 15,147] | 6,030 [3,360 - 16,032] | 6,289 [3,390 - 16,847] | 14,522 [4,801 - 24,401] |
| Mean (SD) | 9,918.17 (7,672.74) | 10,196.57 (9,011.31) | 10,670.43 (9,480.30) | 14,821.73 (10,521.89) | |
| Range | 299 to 38,429 | 299 to 39,430 | 299 to 38,943 | 390 to 39,792 | |
| Future observation | |||||
| - | Median [Q25 - Q75] | 12,868 [6,860 - 18,078] | 13,748 [6,878 - 19,331] | 13,165 [5,988 - 18,548] | 7,798 [2,874 - 14,913] |
| Mean (SD) | 12,865.11 (7,543.50) | 13,470.92 (8,215.96) | 12,913.27 (7,929.17) | 9,167.33 (7,160.81) | |
| Range | 0 to 38,403 | 1 to 39,051 | 0 to 36,654 | 0 to 29,045 | |
| Days in cohort | |||||
| - | Median [Q25 - Q75] | 22 [15 - 29] | 61 [31 - 91] | 61 [31 - 91] | 61 [31 - 91] |
| Mean (SD) | 25.02 (8.00) | 61.65 (25.38) | 62.16 (25.32) | 59.26 (24.79) | |
| Range | 1 to 37 | 2 to 92 | 1 to 91 | 1 to 91 | |
| Medications prior to index date | |||||
| Acetaminophen | N (%) | 1,530 (79.90%) | 357 (76.94%) | 447 (78.56%) | 119 (86.23%) |
| Warfarin | N (%) | 12 (0.63%) | 8 (1.72%) | 11 (1.93%) | 4 (2.90%) |
| Morphine | N (%) | 15 (0.78%) | 1 (0.22%) | 2 (0.35%) | 2 (1.45%) |
| Medications on index date | |||||
| Acetaminophen | N (%) | 773 (40.37%) | 240 (51.72%) | 264 (46.40%) | 90 (65.22%) |
| Morphine | N (%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| Warfarin | N (%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
- Figure 2. Bar chart with proportions of individuals receiving each of the pre-specified list of medicines, devices or procedures, and combinations within a pre-specified time window after diagnosis.
CohortCharacteristics::plotCharacteristics(
result = plot_data,
plotType = "barplot",
colour = "variable_level",
facet = c("cdm_name", "cohort_name")
) 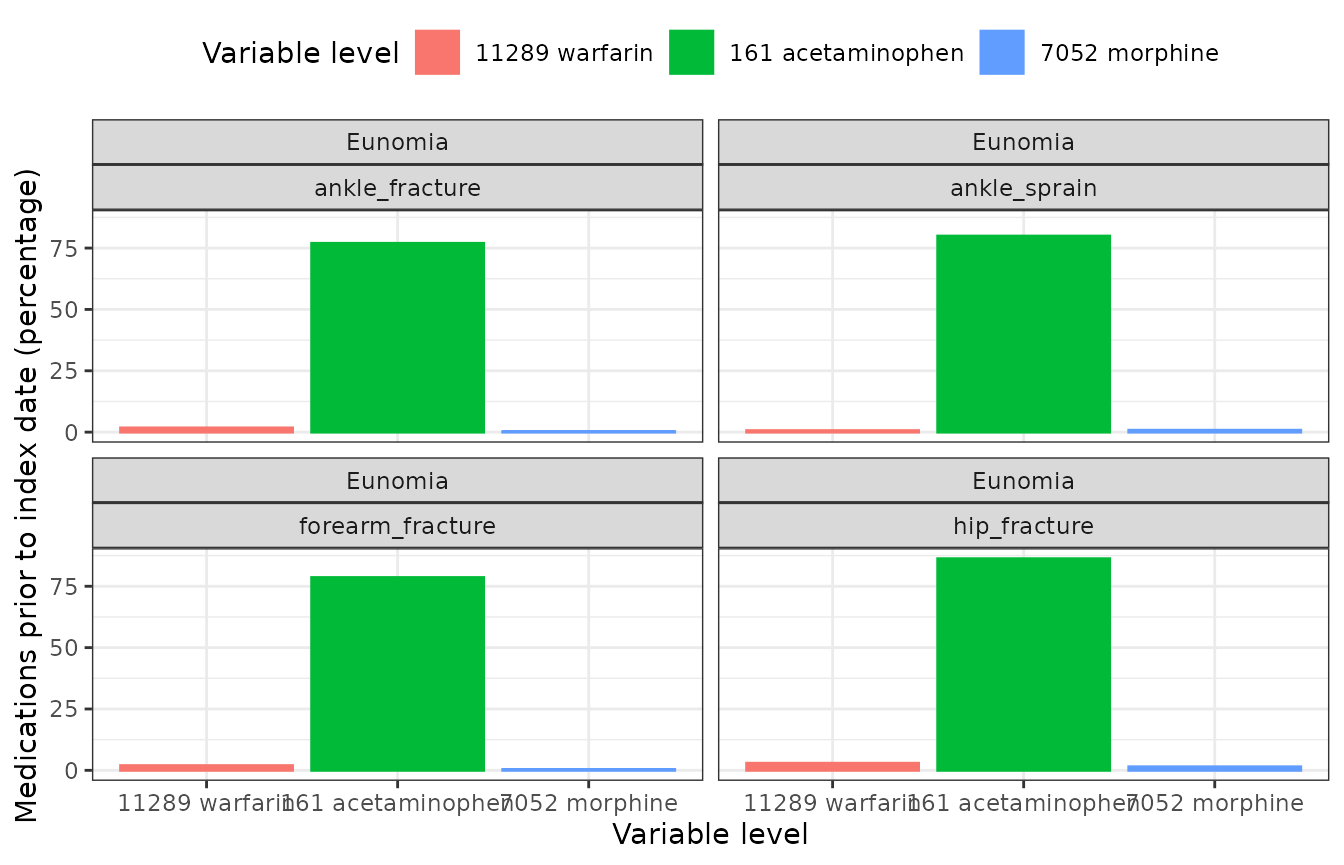 - Figure 3. Sunburst plot depicting treatment patterns
after an index diagnosis.
- Figure 3. Sunburst plot depicting treatment patterns
after an index diagnosis.
TreatmentPatterns::ggSunburst(treatmentPathways = treatmentPathways_data)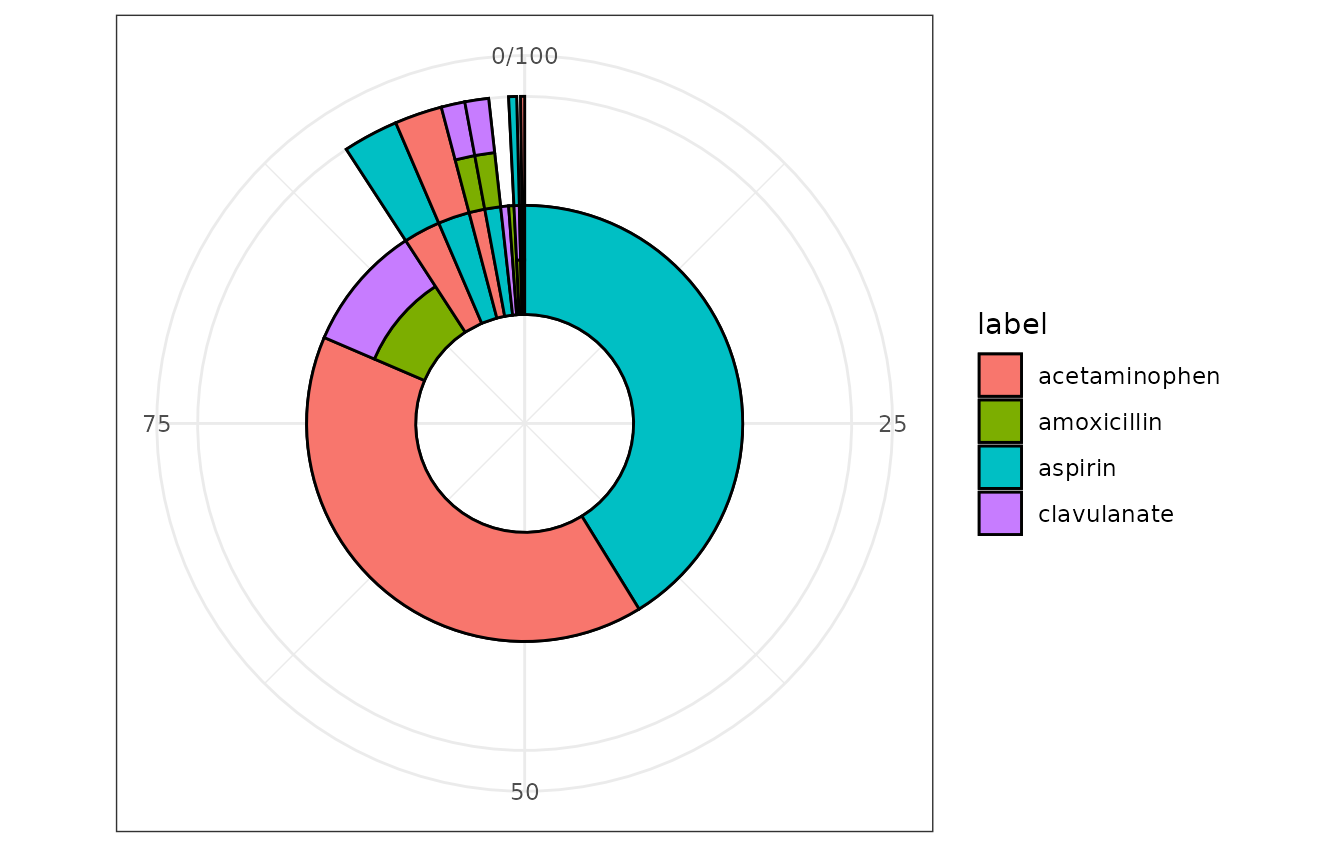 - Figure 4. Sankey diagram depicting treatment
sequences after an index diagnosis.
- Figure 4. Sankey diagram depicting treatment
sequences after an index diagnosis.
TreatmentPatterns::createSankeyDiagram(treatmentPathways = treatmentPathways_data)