For the examples in this vignette glue will be used as
an example. glue version 1.6.2.9000 is included in
the system files of PaRe and is thus accessible even if
these examples are ran offline.
PaRe does fetch some online resources through the
package pak. And by default online stored csv-files in the
PaRe::whiteList data.frame. If no connection can be made,
functions using these methods to reference these online resources will
return NULL.
Whitelist Data Frame
PaRe includes a data frame which contains links to
csv-files to be used in the PaRe::checkDependencies and
PaRe::getDefaultPermittedPackages functions.
By default the data frame contains the following information.
PaRe::whiteList
#> # A tibble: 3 × 4
#> source link package version
#> <chr> <chr> <chr> <chr>
#> 1 tidyverse https://raw.githubusercontent.com/mvankessel-EMC/De… package version
#> 2 darwin https://raw.githubusercontent.com/mvankessel-EMC/De… package version
#> 3 hades https://raw.githubusercontent.com/mvankessel-EMC/De… package versionThe data frame contains 4 columns:
- source: Source name.
- link: Link or path to the csv-file.
- package: Column name in the referenced csv-file that contains the package names.
- version: Column name in the referenced csv-file that contains the package versions.
If you wish to alter the sources in just your R-session, you can either add, remove, or replace individual rows in the whiteList data frame.
sessionWhiteList <- rbind(
whiteList,
list(
source = "dummySession",
link = "some/file.csv",
package = "package",
version = "version"
)
)
sessionWhiteList
#> # A tibble: 4 × 4
#> source link package version
#> <chr> <chr> <chr> <chr>
#> 1 tidyverse https://raw.githubusercontent.com/mvankessel-EMC… package version
#> 2 darwin https://raw.githubusercontent.com/mvankessel-EMC… package version
#> 3 hades https://raw.githubusercontent.com/mvankessel-EMC… package version
#> 4 dummySession some/file.csv package versionIf you wish to make more permanent alterations to the
whiteList data frame, you can edit the whiteList.csv file
in the PaRe system files.
fileWhiteList <- rbind(
read.csv(
system.file(
package = "PaRe",
"whiteList.csv"
)
),
list(
source = "dummyFile",
link = "some/file.csv",
package = "package",
version = "version"
)
)
fileWhiteList
#> source
#> 1 tidyverse
#> 2 darwin
#> 3 hades
#> 4 dummyFile
#> link
#> 1 https://raw.githubusercontent.com/mvankessel-EMC/DependencyReviewerWhitelists/main/tidyverse.csv
#> 2 https://raw.githubusercontent.com/mvankessel-EMC/DependencyReviewerWhitelists/main/darwin.csv
#> 3 https://raw.githubusercontent.com/mvankessel-EMC/DependencyReviewerWhitelists/main/hades.csv
#> 4 some/file.csv
#> package version
#> 1 package version
#> 2 package version
#> 3 package version
#> 4 package version
write.csv(
fileWhiteList,
system.file(
package = "PaRe",
"whiteList.csv"
)
)Dependency Review
Before we start diving into the dependency usage of glue
we should first establish what our dependency white list even looks
like. We can retrieve our full list of whitelisted dependencies buy
calling the getDefaultPermittedPackages function.
getDefaultPermittedPackages
PaRe::getDefaultPermittedPackages(base = TRUE)getDefaultPermittedPackages takes one parameter:
-
base which is set to
TRUEby default. Packages that listed as base packages will be included in the white list.
Setting up a Repository object
# Temp dir to clone repo to
tempDir <- tempdir()
pathToRepo <- file.path(tempDir, "glue")
# Clone IncidencePrevalence to temp dir
git2r::clone(
url = "https://github.com/tidyverse/glue.git",
local_path = pathToRepo
)
repo <- PaRe::Repository$new(path = pathToRepo)#> cloning into 'C:\Users\MVANKE~1\AppData\Local\Temp\RtmpYrand7/glue'...
#> Receiving objects: 1% (63/6253), 63 kb
#> Receiving objects: 11% (688/6253), 592 kb
#> Receiving objects: 21% (1314/6253), 2320 kb
#> Receiving objects: 31% (1939/6253), 2744 kb
#> Receiving objects: 41% (2564/6253), 2856 kb
#> Receiving objects: 51% (3190/6253), 3600 kb
#> Receiving objects: 61% (3815/6253), 3808 kb
#> Receiving objects: 71% (4440/6253), 3920 kb
#> Receiving objects: 81% (5065/6253), 4088 kb
#> Receiving objects: 91% (5691/6253), 4448 kb
#> Receiving objects: 100% (6253/6253), 12641 kb, done.checkDependencies
Now that we know what is included in the white list, we can make our
first step into reviewing glue, which is to ensure the
(suggested) dependencies glue uses are in our white
list.
PaRe::checkDependencies(repo = repo)→ The following are not permitted: covr, microbenchmark, R.utils, rprintf, testthat
→ Please open an issue here: https://github.com/mvankessel-EMC/DependencyReviewerWhitelists/issues| package | version |
|---|---|
| covr | * |
| microbenchmark | * |
| R.utils | * |
| rprintf | * |
| testthat | 3.0.0 |
Not all suggested dependencies are approved. The function prints a message and returns a data frame, containing all packages that are not listed in our white list.
checkDependecies takes two parameters:
- pkgPath which specifies the path to the pacakge.
- dependencyType a vector of character items which specify kinds of imports to look at.
getGraphData
glue depends on (suggested) dependencies. These dependencies in turn import other dependencies, and so on. We can investigate how these recursive dependencies depend on one another, by investigating it as a graph.
graphData <- PaRe::getGraphData(
repo = repo,
packageTypes = c("imports", "suggests")
)We can compute several statistics about our dependency graph
data.frame(
countVertices = length(igraph::V(graphData)),
countEdges = length(igraph::E(graphData)),
meanDegree = round(mean(igraph::degree(graphData)), 2),
meanDistance = round(mean(igraph::distances(graphData)), 2)
)-
countVertices resembles the amount of recursive
dependencies
gluedepends on. - countEdges: are the total amount of imports of all dependencies.
- meanDegree: is the average amount of imports per dependency.
-
meanDistance: is the average amount of dependencies
between
glueand all other recursive dependencies.
We can then plot the graph.
plot(graphData)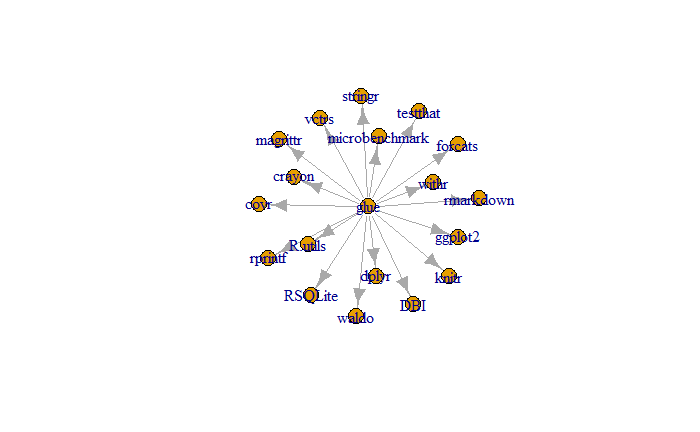
Package wide function use
PaRe allows you to get insight in the function usage in
a package.
summariseFunctionUse
funsUsed <- PaRe::getFunctionUse(repo = repo)
funsUsed#> # A tibble: 367 × 4
#> file line pkg fun
#> <chr> <int> <chr> <chr>
#> 1 color.R 59 base function
#> 2 color.R 59 base parent.frame
#> 3 color.R 60 unknown glue
#> 4 color.R 65 base function
#> 5 color.R 65 base parent.frame
#> 6 color.R 66 unknown glue_data
#> 7 color.R 69 base function
#> 8 color.R 70 base function
#> 9 color.R 70 base parse
#> 10 color.R 70 base tryCatch
#> # ℹ 357 more rowsgetDefinedFunctions
defFuns <- PaRe::getDefinedFunctions(repo = repo)
head(defFuns)#> name lineStart lineEnd nArgs cycloComp fileName
#> 1 glue_col 59 61 3 1 color.R
#> 2 glue_data_col 65 67 4 1 color.R
#> 3 color_transformer 69 99 2 8 color.R
#> 4 intro 67 69 3 1 glue.R
#> 5 glue_data 92 200 6 21 glue.R
#> 6 f 150 176 1 3 glue.RBesides the location of each function being displayed, the number of arguments for each function, and the cyclometic complexity is also included in the result.
PaRe::pkgDiagram(repo = repo) %>%
DiagrammeRsvg::export_svg() %>%
magick::image_read_svg()#> Loading required package: DiagrammeRsvg
#> Loading required package: magick
#> Linking to ImageMagick 6.9.12.98
#> Enabled features: cairo, freetype, fftw, ghostscript, heic, lcms, pango, raw, rsvg, webp
#> Disabled features: fontconfig, x11
Lines of code
PaRe::countPackageLines(repo = repo)#> # A tibble: 1 × 6
#> R cpp o h java sql
#> <int> <int> <int> <int> <int> <int>
#> 1 984 0 0 0 0 0glue contains 1056 lines of R-code.
Linting
#> Warning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
#> dplyr 1.1.0.
#> ℹ Please use `reframe()` instead.
#> ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
#> always returns an ungrouped data frame and adjust accordingly.
#> ℹ The deprecated feature was likely used in the PaRe package.
#> Please report the issue at <https://github.com/darwin-eu-dev/PaRe/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> # A tibble: 2 × 2
#> type pct
#> <chr> <dbl>
#> 1 style 11.9
#> 2 warning 4.07
head(messages)#> filename line_number column_number type
#> 1 color.R 8 81 style
#> 2 color.R 14 81 style
#> 3 color.R 59 1 style
#> 4 color.R 59 81 style
#> 5 color.R 60 3 warning
#> 6 color.R 60 81 style
#> message
#> 1 Lines should not be more than 80 characters. This line is 93 characters.
#> 2 Lines should not be more than 80 characters. This line is 81 characters.
#> 3 Variable and function name style should match camelCase.
#> 4 Lines should not be more than 80 characters. This line is 82 characters.
#> 5 no visible global function definition for 'glue'
#> 6 Lines should not be more than 80 characters. This line is 94 characters.
#> line
#> 1 #' Using the following syntax will apply the function [crayon::blue()] to the text 'foo bar'.
#> 2 #' If you want an expression to be evaluated, simply place that in a normal brace
#> 3 glue_col <- function(..., .envir = parent.frame(), .na = "NA", .literal = FALSE) {
#> 4 glue_col <- function(..., .envir = parent.frame(), .na = "NA", .literal = FALSE) {
#> 5 glue(..., .envir = .envir, .na = .na, .literal = .literal, .transformer = color_transformer)
#> 6 glue(..., .envir = .envir, .na = .na, .literal = .literal, .transformer = color_transformer)
#> linter
#> 1 line_length_linter
#> 2 line_length_linter
#> 3 object_name_linter
#> 4 line_length_linter
#> 5 object_usage_linter
#> 6 line_length_linter