Comparing branches
Maarten van Kessel
2024-11-28
Source:vignettes/articles/ComparingBranches.Rmd
ComparingBranches.Rmdgit package
tempDir <- tempdir()
pathToRepo <- file.path(tempDir, "CDMConnector")
git2r::clone(
url = "https://github.com/darwin-eu/CDMConnector.git",
local_path = pathToRepo
)## cloning into 'C:\Users\MVANKE~1\AppData\Local\Temp\RtmpE9RQLo/CDMConnector'...
## Receiving objects: 1% (86/8529), 124 kb
## Receiving objects: 11% (939/8529), 1436 kb
## Receiving objects: 21% (1792/8529), 4350 kb
## Receiving objects: 31% (2644/8529), 6896 kb
## Receiving objects: 41% (3497/8529), 7064 kb
## Receiving objects: 51% (4350/8529), 8713 kb
## Receiving objects: 61% (5203/8529), 10370 kb
## Receiving objects: 71% (6056/8529), 13115 kb
## Receiving objects: 81% (6909/8529), 16268 kb
## Receiving objects: 91% (7762/8529), 16381 kb
## Receiving objects: 100% (8529/8529), 29661 kb, done.## Local: main C:/Users/mvankessel/AppData/Local/Temp/RtmpE9RQLo/CDMConnector
## Remote: main @ origin (https://github.com/darwin-eu/CDMConnector.git)
## Head: [af064fc] 2024-11-13: V1.6 cran release (#514)Objects
Repository
repo <- PaRe::Repository$new(pathToRepo)Git management
repo$gitCheckout("main")
repo$gitPull()
dev <- repo$clone()
dev$gitCheckout("75e7333cb2d9f8ec7f7a77ae21fb611f8141ef0e")## Switched to: 75e7333cb2d9f8ec7f7a77ae21fb611f8141ef0e## Re-initializingRepository properties
repo$getPath()## [1] "C:\\Users\\mvankessel\\AppData\\Local\\Temp\\RtmpE9RQLo\\CDMConnector"
repo$getName()## [1] "CDMConnector"
repo$getDescription()## Package: CDMConnector
## Title: Connect to an OMOP Common Data Model
## Version: 1.6.0
## Authors@R (parsed):
## * Adam Black <black@ohdsi.org> [aut, cre] (<https://orcid.org/0000-0001-5576-8701>)
## * Artem Gorbachev <artem.gorbachev@odysseusinc.com> [aut]
## * Edward Burn <edward.burn@ndorms.ox.ac.uk> [aut]
## * Marti Catala Sabate <marti.catalasabate@ndorms.ox.ac.uk> [aut]
## Description: Provides tools for working with observational health
## data in the Observational Medical Outcomes Partnership (OMOP) Common
## Data Model format with a pipe friendly syntax. Common data model
## database table references are stored in a single compound object along
## with metadata.
## License: Apache License (>= 2)
## URL: https://darwin-eu.github.io/CDMConnector/,
## https://github.com/darwin-eu/CDMConnector
## BugReports: https://github.com/darwin-eu/CDMConnector/issues
## Depends:
## R (>= 4.0)
## Imports:
## checkmate,
## cli,
## DBI (>= 0.3.0),
## dbplyr (>= 2.5.0),
## dplyr,
## fs,
## generics,
## glue,
## jsonlite,
## lifecycle,
## methods,
## omopgenerics (>= 0.1.2),
## purrr,
## readr,
## rlang,
## stringi,
## stringr,
## tidyr,
## tidyselect,
## waldo,
## withr
## Suggests:
## bigrquery,
## CirceR,
## clock,
## covr,
## DatabaseConnector,
## duckdb,
## ggplot2,
## knitr,
## lubridate,
## odbc,
## palmerpenguins,
## pool,
## rJava,
## rmarkdown,
## RPostgres,
## RSQLite,
## snakecase,
## SqlRender,
## testthat (>= 3.0.0),
## tibble,
## tictoc
## Enhances:
## arrow
## VignetteBuilder:
## knitr
## Config/testthat/edition: 3
## Config/testthat/parallel: false
## Encoding: UTF-8
## Roxygen: list(markdown = TRUE)
## RoxygenNote: 7.3.2
## Collate:
## 'CDMConnector-package.R'
## 'Eunomia.R'
## 'benchmarkCDMConnector.R'
## 'cdm.R'
## 'cdmSubset.R'
## 'cdm_from_environment.R'
## 'cohortTransformations.R'
## 'cohort_ddl.R'
## 'compute.R'
## 'copy_cdm_to.R'
## 'dateadd.R'
## 'dbSource.R'
## 'reexports-omopgenerics.R'
## 'generateCohortSet.R'
## 'generateConceptCohortSet.R'
## 'summariseQuantile.R'
## 'utils.R'
## 'validate.R'
## 'zzz-deprecated.R'File
files <- repo$getFiles()
files## $R
## $R[[1]]
## <File> <Code> <R6>
## Name: benchmarkCDMConnector.R
## # Lines: 138
## $R[[2]]
## <File> <Code> <R6>
## Name: cdm.R
## # Lines: 971
## $R[[3]]
## <File> <Code> <R6>
## Name: cdm_from_environment.R
## # Lines: 196
## $R[[4]]
## <File> <Code> <R6>
## Name: CDMConnector-package.R
## # Lines: 14
## $R[[5]]
## <File> <Code> <R6>
## Name: cdmSubset.R
## # Lines: 520
## $R[[6]]
## <File> <Code> <R6>
## Name: cohort_ddl.R
## # Lines: 126
## $R[[7]]
## <File> <Code> <R6>
## Name: cohortTransformations.R
## # Lines: 380
## $R[[8]]
## <File> <Code> <R6>
## Name: compute.R
## # Lines: 360
## $R[[9]]
## <File> <Code> <R6>
## Name: copy_cdm_to.R
## # Lines: 103
## $R[[10]]
## <File> <Code> <R6>
## Name: dateadd.R
## # Lines: 260
## $R[[11]]
## <File> <Code> <R6>
## Name: dbSource.R
## # Lines: 281
## $R[[12]]
## <File> <Code> <R6>
## Name: Eunomia.R
## # Lines: 406
## $R[[13]]
## <File> <Code> <R6>
## Name: generateCohortSet.R
## # Lines: 983
## $R[[14]]
## <File> <Code> <R6>
## Name: generateConceptCohortSet.R
## # Lines: 452
## $R[[15]]
## <File> <Code> <R6>
## Name: reexports-omopgenerics.R
## # Lines: 95
## $R[[16]]
## <File> <Code> <R6>
## Name: summariseQuantile.R
## # Lines: 146
## $R[[17]]
## <File> <Code> <R6>
## Name: utils.R
## # Lines: 280
## $R[[18]]
## <File> <Code> <R6>
## Name: validate.R
## # Lines: 246
## $R[[19]]
## <File> <Code> <R6>
## Name: zzz-deprecated.R
## # Lines: 121
##
## $cpp
## list()
##
## $o
## list()
##
## $h
## list()
##
## $java
## list()
##
## $sql
## list()
file <- files$R[[1]]
file## <File> <Code> <R6>
## Name: benchmarkCDMConnector.R
## # Lines: 138File properties
file$getName()## [1] "benchmarkCDMConnector.R"
file$getNLines()## [1] 138
file$getType()## [1] "R"
head(file$getLines())## [1] "# Copyright 2024 DARWIN EU®"
## [2] "#"
## [3] "# This file is part of CDMConnector"
## [4] "#"
## [5] "# Licensed under the Apache License, Version 2.0 (the \"License\");"
## [6] "# you may not use this file except in compliance with the License."
tail(file$getLines())## [1] " dplyr::count() %>%"
## [2] " dplyr::pull())"
## [3] ""
## [4] ""
## [5] " return(timings)"
## [6] "}"
file$getFunctionTable()## name lineStart lineEnd nArgs cycloComp
## 1 benchmarkCDMConnector 33 138 1 1Function
functions <- file$getFunctions()
functions## [[1]]
## <Function> <Code> <R6>
## Name: benchmarkCDMConnector
## # Lines: 106## <Function> <Code> <R6>
## Name: benchmarkCDMConnector
## # Lines: 106Function properties
fun$getName()## [1] "benchmarkCDMConnector"
fun$getNLines()## [1] 106
fun$getFunction()## name lineStart lineEnd nArgs cycloComp
## 1 benchmarkCDMConnector 33 138 1 1
head(fun$getLines())## [1] "benchmarkCDMConnector <- function(cdm) {"
## [2] ""
## [3] " checkmate::assertClass(cdm, \"cdm_reference\")"
## [4] ""
## [5] " # will add timings to list"
## [6] " timings <- list()"
tail(fun$getLines())## [1] " dplyr::count() %>%"
## [2] " dplyr::pull())"
## [3] ""
## [4] ""
## [5] " return(timings)"
## [6] "}"ComPaRing main to develop
dplyr::bind_rows(
PaRe::countPackageLines(repo) %>% mutate(branch = "main"),
PaRe::countPackageLines(dev) %>% mutate(branch = "V0.1.0")
)## # A tibble: 2 × 7
## R cpp o h java sql branch
## <int> <int> <int> <int> <int> <int> <chr>
## 1 6078 0 0 0 0 0 main
## 2 2628 0 0 0 0 0 V0.1.0
PaRe::pkgDiagram(repo)
PaRe::pkgDiagram(dev)
mainFunUse <- PaRe::getFunctionUse(repo) %>%
mutate(branch = "main")
devFunUse <- PaRe::getFunctionUse(dev) %>%
mutate(branch = "V0.1.0")
pkgUse <- dplyr::bind_rows(
mainFunUse,
devFunUse
) %>%
group_by(pkg, branch) %>%
tally()
library(ggplot2)
ggplot(data = pkgUse, mapping = aes(x = pkg, y = n, fill = branch)) +
geom_bar(stat = "identity", position = "dodge") +
theme_bw() +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1))
mainFuns <- PaRe::getDefinedFunctions(repo)
devFuns <- PaRe::getDefinedFunctions(dev)
defFuns <- bind_rows(
mainFuns %>% mutate(branch = "main"),
devFuns %>% mutate(branch = "V0.1.0")
)
ggplot(data = defFuns, mapping = aes(y = cycloComp, x = name, fill = branch)) +
geom_bar(stat = "identity", position = "dodge") +
theme_bw() +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1)) +
labs(title = "Cyclomatic Complexity per Function", x = "Function", y = "Cyclomatic Complexity")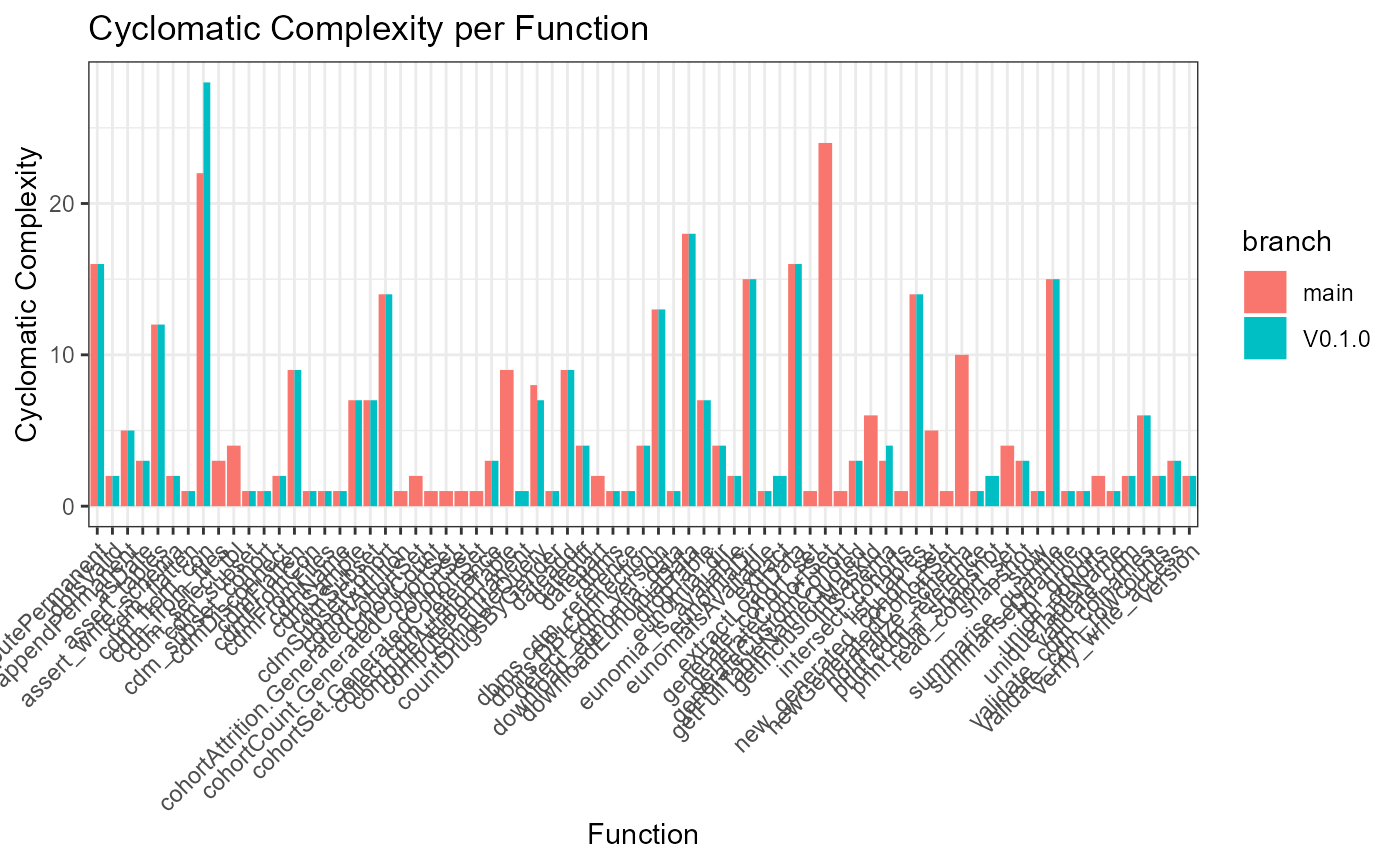
ggplot(data = defFuns, mapping = aes(y = lineEnd - lineStart + 1, x = name, fill = branch)) +
geom_bar(stat = "identity", position = "dodge") +
theme_bw() +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1)) +
labs(title = "Number of Lines per Function", y = "Number of Lines", x = "Function")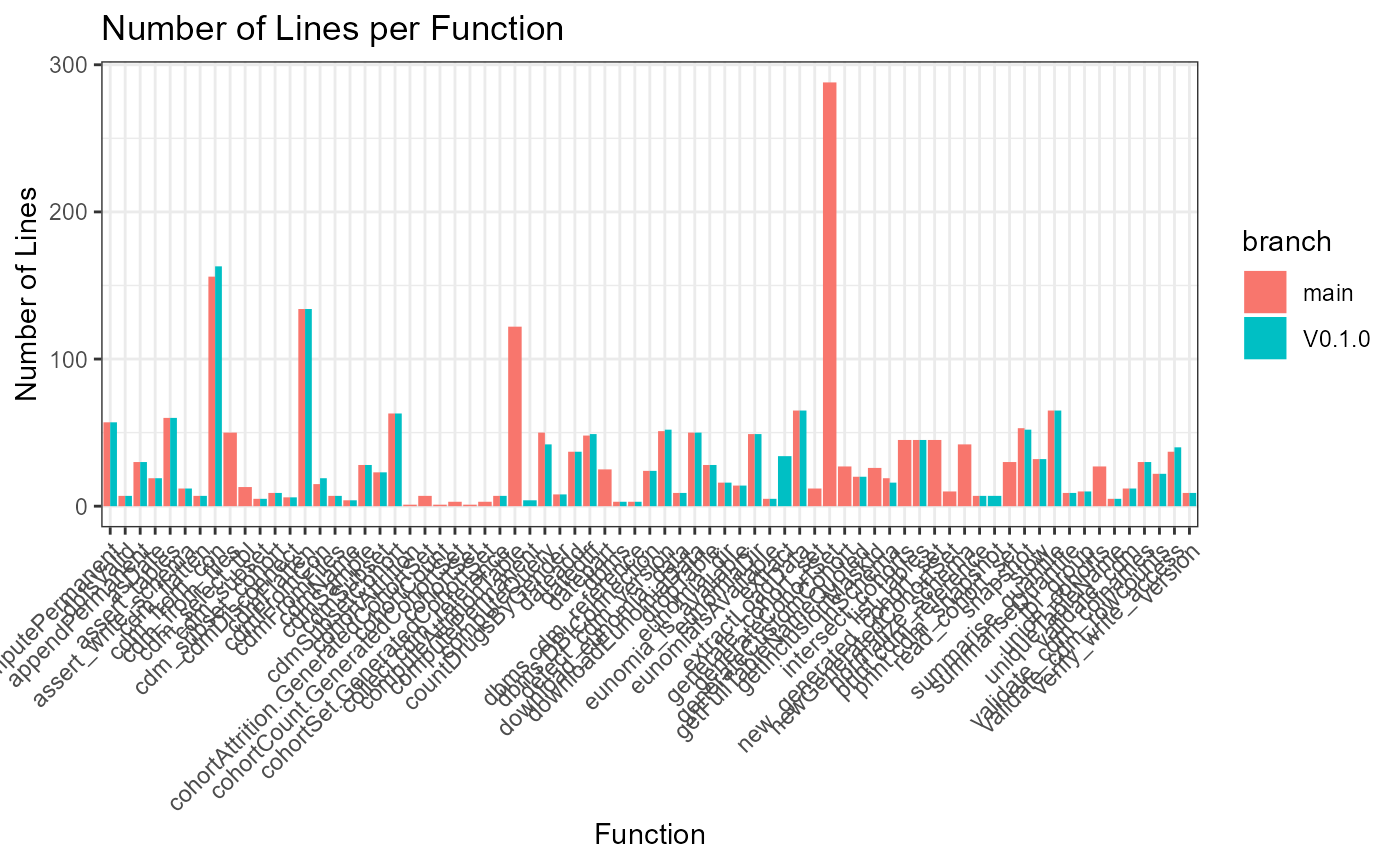
effectiveCode <- defFuns %>%
group_by(fileName, branch) %>%
summarise(n = sum(lineEnd - lineStart + 1)) %>%
ungroup()## `summarise()` has grouped output by 'fileName'. You can override using the
## `.groups` argument.
ggplot(data = effectiveCode, mapping = aes(y = n, x = fileName, fill = branch)) +
geom_bar(stat = "identity", position = "dodge") +
theme_bw() +
theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust = 1)) +
labs(title = "Functional code increase", y = "Number of Lines", x = "File")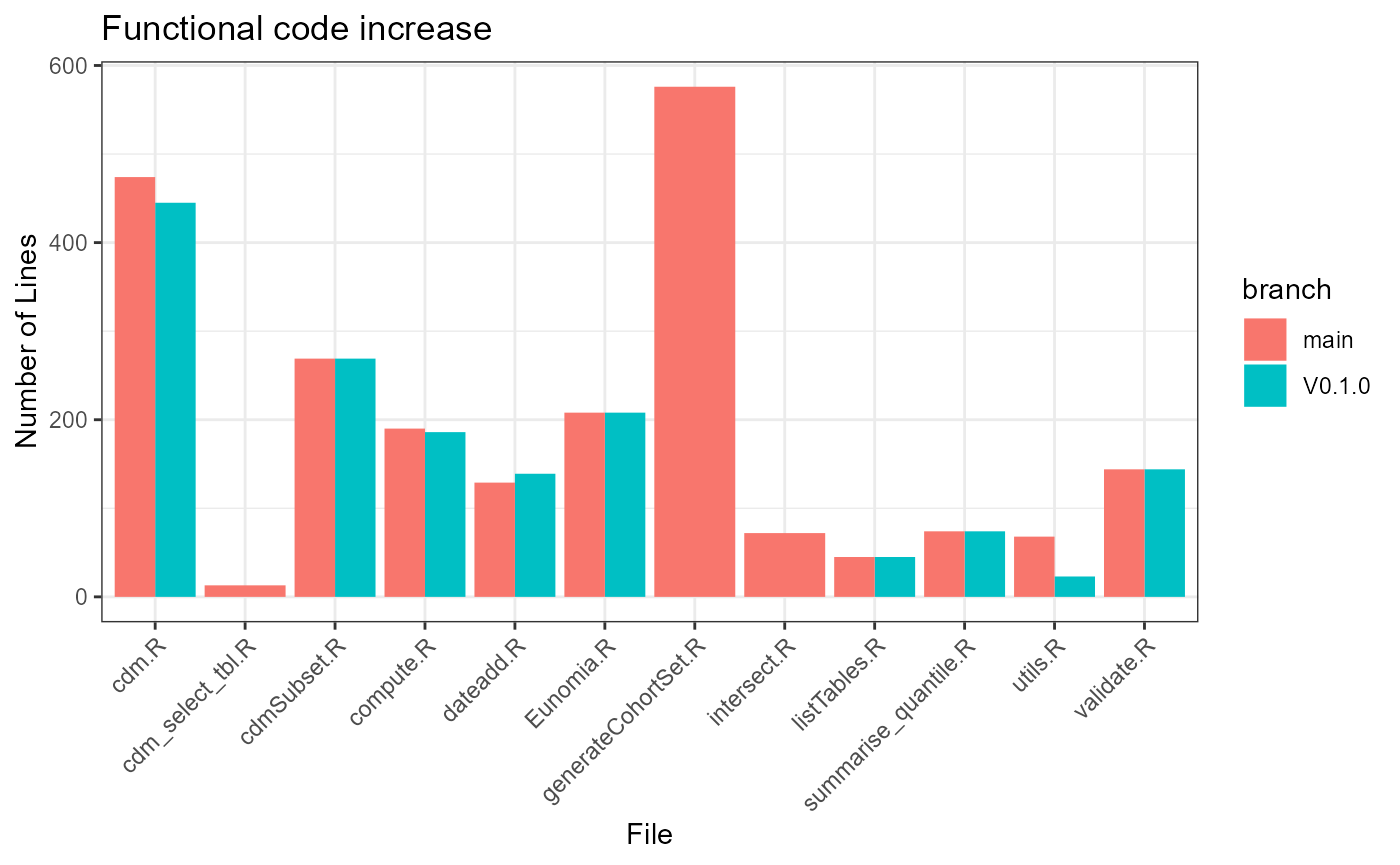
mainGraph <- PaRe::getGraphData(repo)## ✔ Updated metadata database: 5.54 MB in 15 files.
## ℹ Updating metadata database✔ Updating metadata database ... done## Warning in getParDeps(pkgs = deps, nThreads = nThreads): Could not fetch
## dependencies for package: `methods`
devGraph <- PaRe::getGraphData(dev)## Warning in getParDeps(pkgs = deps, nThreads = nThreads): Could not fetch
## dependencies for package: `methods`##
## Attaching package: 'igraph'
##
## The following objects are masked from 'package:dplyr':
##
## as_data_frame, groups, union
##
## The following objects are masked from 'package:stats':
##
## decompose, spectrum
##
## The following object is masked from 'package:base':
##
## union
data.frame(
branch = c("main", "V0.1.0"),
countVertices = c(length(igraph::V(mainGraph)), length(igraph::V(devGraph))),
countEdges = c(length(igraph::E(mainGraph)), length(igraph::E(devGraph))),
meanDegree = c(round(mean(igraph::degree(mainGraph)), 2), round(mean(igraph::degree(devGraph)), 2)),
meanDistance = c(round(mean(igraph::distances(mainGraph)), 2), round(mean(igraph::distances(devGraph)), 2))
)## branch countVertices countEdges meanDegree meanDistance
## 1 main 45 526 23.38 2.15
## 2 V0.1.0 43 348 16.19 2.15## filename line_number column_number type
## 1 benchmarkCDMConnector.R 110 81 style
## 2 cdm.R 21 81 style
## 3 cdm.R 23 81 style
## 4 cdm.R 25 81 style
## 5 cdm.R 27 81 style
## 6 cdm.R 30 81 style
## message
## 1 Lines should not be more than 80 characters. This line is 82 characters.
## 2 Lines should not be more than 80 characters. This line is 89 characters.
## 3 Lines should not be more than 80 characters. This line is 91 characters.
## 4 Lines should not be more than 80 characters. This line is 92 characters.
## 5 Lines should not be more than 80 characters. This line is 85 characters.
## 6 Lines should not be more than 80 characters. This line is 85 characters.
## line
## 1 task <- "summary of observation period start and end dates by gender concept id"
## 2 #' @param cdm_schema,cdmSchema The schema where the OMOP CDM tables are located. Defaults
## 3 #' @param write_schema,writeSchema An optional schema in the CDM database that the user has
## 4 #' @param cohort_tables,cohortTables A character vector listing the cohort table names to be
## 5 #' @param cdm_version,cdmVersion The version of the OMOP CDM: "5.3" (default), "5.4",
## 6 #' @param cdm_name,cdmName The name of the CDM. If NULL (default) the cdm_source_name
## linter
## 1 line_length_linter
## 2 line_length_linter
## 3 line_length_linter
## 4 line_length_linter
## 5 line_length_linter
## 6 line_length_linter
bind_rows(
PaRe::lintScore(repo, messages) %>% mutate(branch = "main"),
PaRe::lintScore(dev, messages) %>% mutate(branch = "v0.1.0")
)## # A tibble: 4 × 3
## type pct branch
## <chr> <dbl> <chr>
## 1 style 14.9 main
## 2 warning 3.37 main
## 3 style 34.4 v0.1.0
## 4 warning 7.8 v0.1.0