Subject ID conversion
As of TreatmentPatterns 2.7.0 TreatmentPatterns will
assign it’s own pseudo subject ID’s. This is due to subject ID’s in the
database may be 64 bit integers, or may contain letters. This can lead
to strange edge cases where ID’s get rounded, and results of different
individuals will be grouped together.
The real ID’s that exist in the database will be stored as a
character(1) in the cohortTable table, in the
patient level result object from computePathways()
In the following snipit of code we will show how to relate the internal ID’s to the ID’s existing in the database:
##
## Attaching package: 'dplyr'## The following objects are masked from 'package:stats':
##
## filter, lag## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomia_dir())
cdm <- cdmFromCon(con, cdmSchema = "main", writeSchema = "main")
cohortSet <- readCohortSet(
path = system.file(package = "TreatmentPatterns", "exampleCohorts")
)
cdm <- generateCohortSet(
cdm = cdm,
cohortSet = cohortSet,
name = "cohort_table"
)## ℹ Generating 8 cohorts## ℹ Generating cohort (1/8) - acetaminophen✔ Generating cohort (1/8) - acetaminophen [268ms]
## ℹ Generating cohort (2/8) - amoxicillin✔ Generating cohort (2/8) - amoxicillin [177ms]
## ℹ Generating cohort (3/8) - aspirin✔ Generating cohort (3/8) - aspirin [166ms]
## ℹ Generating cohort (4/8) - clavulanate✔ Generating cohort (4/8) - clavulanate [181ms]
## ℹ Generating cohort (5/8) - death✔ Generating cohort (5/8) - death [104ms]
## ℹ Generating cohort (6/8) - doxylamine✔ Generating cohort (6/8) - doxylamine [158ms]
## ℹ Generating cohort (7/8) - penicillinv✔ Generating cohort (7/8) - penicillinv [161ms]
## ℹ Generating cohort (8/8) - viralsinusitis✔ Generating cohort (8/8) - viralsinusitis [245ms]## Warning: ! 5 casted column in cohort_table (cohort_attrition) as do not match expected
## column type:
## • `number_records` from numeric to integer
## • `number_subjects` from numeric to integer
## • `reason_id` from numeric to integer
## • `excluded_records` from numeric to integer
## • `excluded_subjects` from numeric to integer## Warning: ! 1 column in cohort_table do not match expected column type:
## • `subject_id` is numeric but expected integer
# convert subject_id to subject_id + 1e16 as 64 bit integer
cdm$cohort_table <- cdm$cohort_table %>%
mutate(subject_id = bit64::as.integer64(.data$subject_id + 1e16)) %>%
compute()
cdm$person <- cdm$person %>%
mutate(person_id = bit64::as.integer64(.data$person_id + 1e16)) %>%
compute()
cohorts <- cohortSet %>%
# Remove 'cohort' and 'json' columns
select(-"cohort", -"json") %>%
mutate(type = c("event", "event", "event", "event", "exit", "event", "event", "target")) %>%
rename(
cohortId = "cohort_definition_id",
cohortName = "cohort_name",
) %>%
select("cohortId", "cohortName", "type")
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm
)## -- Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## Records: 14041
## Subjects: 2693
## -- Removing records < minEraDuration (0)
## Records: 11386
## Subjects: 2159
## -- Removing events where index date < target index date + indexDateOffset (0)
## Records: 8381
## Subjects: 2159
## -- splitEventCohorts
## Records: 8366
## Subjects: 2144
## -- eraCollapse (30)
## Records: 8366
## Subjects: 2144
## -- Iteration 1: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 558
## Subjects: 512
## -- Iteration 2: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 554
## Subjects: 512
## -- After Combination
## Records: 554
## Subjects: 512
## -- filterTreatments (First)
## Records: 553
## Subjects: 512
## -- treatment construction done
## Records: 553
## Subjects: 512
outputEnv$treatmentHistory %>%
inner_join(outputEnv$cohortTable, join_by(personId == personId)) %>%
select("personId", "subject_id_origin") %>%
head()## # Source: SQL [6 x 2]
## # Database: sqlite 3.46.0 [C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ\file79f820ad2da0.sqlite]
## personId subject_id_origin
## <dbl> <chr>
## 1 324 10000000000000836
## 2 324 10000000000000836
## 3 324 10000000000000836
## 4 324 10000000000000836
## 5 426 10000000000005148
## 6 426 10000000000005148
Andromeda::close(outputEnv)
DBI::dbDisconnect(con, shutdown = TRUE)
periodPriorToIndex to indexDateOffset
The periodPriorToIndex parameter has been renamed to
indexDateOffset. It was already possible to use the
parameter as an offset, with negative numbers. But this change
solidifies this use-case.
Note that the logic is not inversed. If you want to look back for 30
days prior to the index date previously you would set
periodPriorToIndex = 30 which now equals
indexDateOffset = -30.
library(TreatmentPatterns)
library(CDMConnector)
library(dplyr)
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomia_dir())
cdm <- cdmFromCon(con, cdmSchema = "main", writeSchema = "main")
cohortSet <- readCohortSet(
path = system.file(package = "TreatmentPatterns", "exampleCohorts")
)
cdm <- generateCohortSet(
cdm = cdm,
cohortSet = cohortSet,
name = "cohort_table"
)## ℹ Generating 8 cohorts## ℹ Generating cohort (1/8) - acetaminophen✔ Generating cohort (1/8) - acetaminophen [169ms]
## ℹ Generating cohort (2/8) - amoxicillin✔ Generating cohort (2/8) - amoxicillin [158ms]
## ℹ Generating cohort (3/8) - aspirin✔ Generating cohort (3/8) - aspirin [164ms]
## ℹ Generating cohort (4/8) - clavulanate✔ Generating cohort (4/8) - clavulanate [141ms]
## ℹ Generating cohort (5/8) - death✔ Generating cohort (5/8) - death [98ms]
## ℹ Generating cohort (6/8) - doxylamine✔ Generating cohort (6/8) - doxylamine [143ms]
## ℹ Generating cohort (7/8) - penicillinv✔ Generating cohort (7/8) - penicillinv [157ms]
## ℹ Generating cohort (8/8) - viralsinusitis✔ Generating cohort (8/8) - viralsinusitis [243ms]## Warning: ! 5 casted column in cohort_table (cohort_attrition) as do not match expected
## column type:
## • `number_records` from numeric to integer
## • `number_subjects` from numeric to integer
## • `reason_id` from numeric to integer
## • `excluded_records` from numeric to integer
## • `excluded_subjects` from numeric to integer## Warning: ! 1 column in cohort_table do not match expected column type:
## • `subject_id` is numeric but expected integer
# convert subject_id to subject_id + 1e16 as 64 bit integer
cdm$cohort_table <- cdm$cohort_table %>%
mutate(subject_id = bit64::as.integer64(.data$subject_id + 1e16)) %>%
compute()
cdm$person <- cdm$person %>%
mutate(person_id = bit64::as.integer64(.data$person_id + 1e16)) %>%
compute()
cohorts <- cohortSet %>%
# Remove 'cohort' and 'json' columns
select(-"cohort", -"json") %>%
mutate(type = c("event", "event", "event", "event", "exit", "event", "event", "target")) %>%
rename(
cohortId = "cohort_definition_id",
cohortName = "cohort_name",
) %>%
select("cohortId", "cohortName", "type")
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm,
# Look back (index date - 30 days)
indexDateOffset = -30
)## -- Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## Records: 14041
## Subjects: 2693
## -- Removing records < minEraDuration (0)
## Records: 11386
## Subjects: 2159
## -- Removing events where index date < target index date + indexDateOffset (-30)
## Records: 8381
## Subjects: 2159
## -- splitEventCohorts
## Records: 8366
## Subjects: 2144
## -- eraCollapse (30)
## Records: 8366
## Subjects: 2144
## -- Iteration 1: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 558
## Subjects: 512
## -- Iteration 2: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 554
## Subjects: 512
## -- After Combination
## Records: 554
## Subjects: 512
## -- filterTreatments (First)
## Records: 553
## Subjects: 512
## -- treatment construction done
## Records: 553
## Subjects: 512
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm,
# Look ahead (index date + 30 days)
indexDateOffset = 30
)## -- Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## Records: 14041
## Subjects: 2693
## -- Removing records < minEraDuration (0)
## Records: 11386
## Subjects: 2159
## -- Removing events where index date < target index date + indexDateOffset (30)
## Records: 6349
## Subjects: 2159
## -- splitEventCohorts
## Records: 6267
## Subjects: 2077
## -- eraCollapse (30)
## Records: 6267
## Subjects: 2077
## -- Iteration 1: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 362
## Subjects: 341
## -- Iteration 2: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 359
## Subjects: 341
## -- After Combination
## Records: 359
## Subjects: 341
## -- filterTreatments (First)
## Records: 358
## Subjects: 341
## -- treatment construction done
## Records: 358
## Subjects: 341Empty data
In previous versions TreatmentPatterns, TreatmentPatterns would throw
an ambiguous error from dplyr if the target
was empty, wasn’t assigned or other related issues.
Now TreatmentPatterns throws more informative errors and warnings when something like this is detected.
library(TreatmentPatterns)
library(CDMConnector)
library(dplyr)
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomia_dir())
cdm <- cdmFromCon(con, cdmSchema = "main", writeSchema = "main")
cohortSet <- readCohortSet(
path = system.file(package = "TreatmentPatterns", "exampleCohorts")
)
cdm <- generateCohortSet(
cdm = cdm,
cohortSet = cohortSet,
name = "cohort_table"
)## ℹ Generating 8 cohorts## ℹ Generating cohort (1/8) - acetaminophen✔ Generating cohort (1/8) - acetaminophen [152ms]
## ℹ Generating cohort (2/8) - amoxicillin✔ Generating cohort (2/8) - amoxicillin [158ms]
## ℹ Generating cohort (3/8) - aspirin✔ Generating cohort (3/8) - aspirin [136ms]
## ℹ Generating cohort (4/8) - clavulanate✔ Generating cohort (4/8) - clavulanate [143ms]
## ℹ Generating cohort (5/8) - death✔ Generating cohort (5/8) - death [100ms]
## ℹ Generating cohort (6/8) - doxylamine✔ Generating cohort (6/8) - doxylamine [151ms]
## ℹ Generating cohort (7/8) - penicillinv✔ Generating cohort (7/8) - penicillinv [142ms]
## ℹ Generating cohort (8/8) - viralsinusitis✔ Generating cohort (8/8) - viralsinusitis [226ms]## Warning: ! 5 casted column in cohort_table (cohort_attrition) as do not match expected
## column type:
## • `number_records` from numeric to integer
## • `number_subjects` from numeric to integer
## • `reason_id` from numeric to integer
## • `excluded_records` from numeric to integer
## • `excluded_subjects` from numeric to integer## Warning: ! 1 column in cohort_table do not match expected column type:
## • `subject_id` is numeric but expected integer
# convert subject_id to subject_id + 1e16 as 64 bit integer
cdm$cohort_table <- cdm$cohort_table %>%
mutate(subject_id = bit64::as.integer64(.data$subject_id + 1e16)) %>%
compute()
cdm$person <- cdm$person %>%
mutate(person_id = bit64::as.integer64(.data$person_id + 1e16)) %>%
compute()
cohorts <- cohortSet %>%
# Remove 'cohort' and 'json' columns
select(-"cohort", -"json") %>%
mutate(type = c("event", "event", "event", "event", "exit", "event", "event", "target")) %>%
rename(
cohortId = "cohort_definition_id",
cohortName = "cohort_name",
) %>%
select("cohortId", "cohortName", "type")
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm,
# Set minEraDuration, minPostCombinationWindow, and combinationWindow to
# unreasonably large values, so no record qualifies.
minEraDuration = 9999999,
minPostCombinationDuration = 9999999,
combinationWindow = 9999999
)## -- Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## Records: 14041
## Subjects: 2693
## -- Removing records < minEraDuration (9999999)
## Records: 0
## Subjects: 0
## -- Removing events where index date < target index date + indexDateOffset (0)
## Records: 0
## Subjects: 0## Warning in constructPathways(settings = args, andromeda = andromeda): No cases
## found. Generating empty treatmentHistory table.## -- treatment construction done
## Records: 0
## Subjects: 0
cohorts <- cohortSet %>%
# Remove 'cohort' and 'json' columns
select(-"cohort", -"json") %>%
# Only set "event" types
mutate(type = c("event", "event", "event", "event", "event", "event", "event", "event")) %>%
rename(
cohortId = "cohort_definition_id",
cohortName = "cohort_name",
) %>%
select("cohortId", "cohortName", "type")
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm
)## Error in validateComputePathways(): No 'target' cohort specified in `cohorts`.
summaryStatsTherapyDuration.csv to
summaryEventDuration.csv
The summaryStatsTherapyDuration.csv has been renamed to
summaryEventDuration.csv. The file now fully focuses on the
duration of events, across different levels in the pathways, and
overall.
library(TreatmentPatterns)
library(CDMConnector)
library(dplyr)
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomia_dir())
cdm <- cdmFromCon(con, cdmSchema = "main", writeSchema = "main")
cohortSet <- readCohortSet(
path = system.file(package = "TreatmentPatterns", "exampleCohorts")
)
cdm <- generateCohortSet(
cdm = cdm,
cohortSet = cohortSet,
name = "cohort_table"
)## ℹ Generating 8 cohorts## ℹ Generating cohort (1/8) - acetaminophen✔ Generating cohort (1/8) - acetaminophen [150ms]
## ℹ Generating cohort (2/8) - amoxicillin✔ Generating cohort (2/8) - amoxicillin [146ms]
## ℹ Generating cohort (3/8) - aspirin✔ Generating cohort (3/8) - aspirin [150ms]
## ℹ Generating cohort (4/8) - clavulanate✔ Generating cohort (4/8) - clavulanate [147ms]
## ℹ Generating cohort (5/8) - death✔ Generating cohort (5/8) - death [105ms]
## ℹ Generating cohort (6/8) - doxylamine✔ Generating cohort (6/8) - doxylamine [147ms]
## ℹ Generating cohort (7/8) - penicillinv✔ Generating cohort (7/8) - penicillinv [157ms]
## ℹ Generating cohort (8/8) - viralsinusitis✔ Generating cohort (8/8) - viralsinusitis [218ms]## Warning: ! 5 casted column in cohort_table (cohort_attrition) as do not match expected
## column type:
## • `number_records` from numeric to integer
## • `number_subjects` from numeric to integer
## • `reason_id` from numeric to integer
## • `excluded_records` from numeric to integer
## • `excluded_subjects` from numeric to integer## Warning: ! 1 column in cohort_table do not match expected column type:
## • `subject_id` is numeric but expected integer
# convert subject_id to subject_id + 1e16 as 64 bit integer
cdm$cohort_table <- cdm$cohort_table %>%
mutate(subject_id = bit64::as.integer64(.data$subject_id + 1e16)) %>%
compute()
cdm$person <- cdm$person %>%
mutate(person_id = bit64::as.integer64(.data$person_id + 1e16)) %>%
compute()
cohorts <- cohortSet %>%
# Remove 'cohort' and 'json' columns
select(-"cohort", -"json") %>%
mutate(type = c("event", "event", "event", "event", "exit", "event", "event", "target")) %>%
rename(
cohortId = "cohort_definition_id",
cohortName = "cohort_name",
) %>%
select("cohortId", "cohortName", "type")
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm
)## -- Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## Records: 14041
## Subjects: 2693
## -- Removing records < minEraDuration (0)
## Records: 11386
## Subjects: 2159
## -- Removing events where index date < target index date + indexDateOffset (0)
## Records: 8381
## Subjects: 2159
## -- splitEventCohorts
## Records: 8366
## Subjects: 2144
## -- eraCollapse (30)
## Records: 8366
## Subjects: 2144
## -- Iteration 1: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 558
## Subjects: 512
## -- Iteration 2: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 554
## Subjects: 512
## -- After Combination
## Records: 554
## Subjects: 512
## -- filterTreatments (First)
## Records: 553
## Subjects: 512
## -- treatment construction done
## Records: 553
## Subjects: 512## Writing attrition to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/attrition.csv
## Writing metadata to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/metadata.csv
## Writing treatmentPathways to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/treatmentPathways.csv
## Censoring 1219 pathways with a frequency <5 to 5.
## Writing summaryStatsTherapyDuration to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/summaryEventDuration.csv
## Writing countsYearPath to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/countsYear.csv
## Writing countsAgePath to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/countsAge.csv
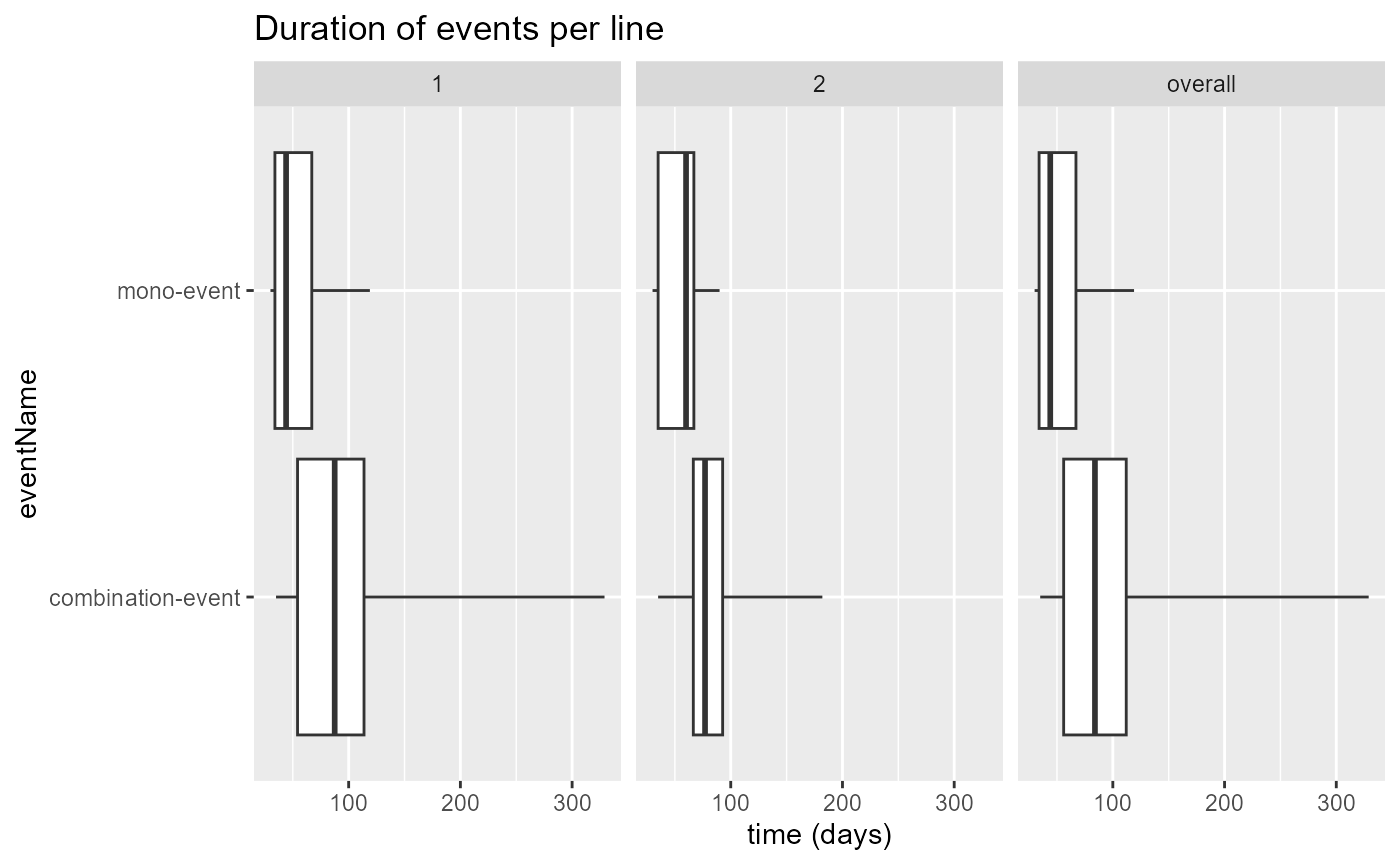
## Writing countsSexPath to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/countsSex.csv## eventName min Q1 median Q2 max average sd count line
## 1 combination-event 35 56.00 84.0 112.00 329 90.23438 48.59575 1711 overall
## 2 mono-event 30 34.00 44.0 67.00 119 53.97751 23.84533 489 overall
## 3 combination-event 35 54.25 87.5 113.75 329 91.94231 50.85790 1699 1
## 4 combination-event 35 66.50 77.0 92.75 182 82.83333 38.20479 12 2
## 5 mono-event 30 34.00 44.0 67.00 119 53.90217 23.88975 460 1
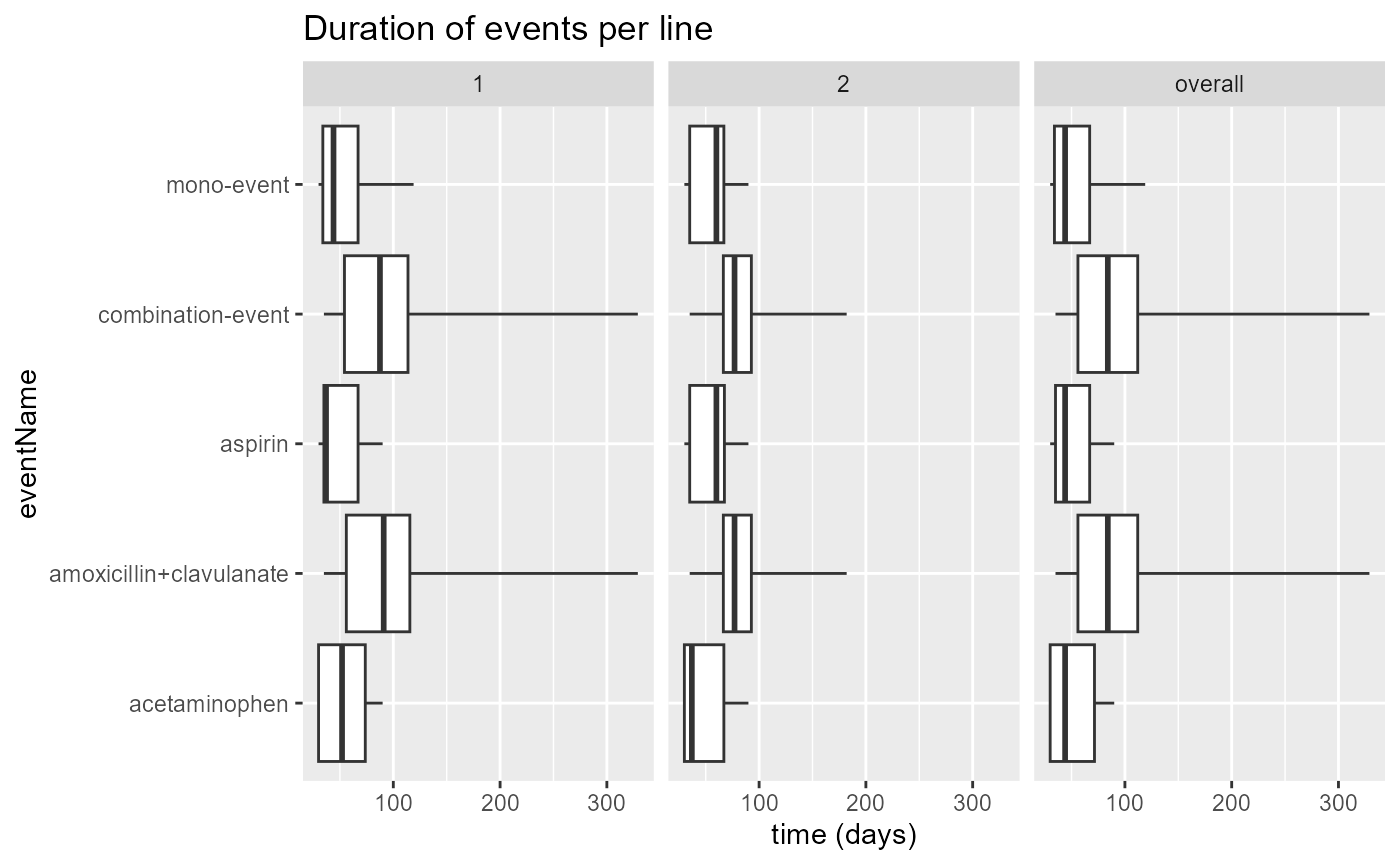
## 6 mono-event 30 35.00 60.0 67.00 90 55.17241 23.50694 29 2Event duration with plotEventDuration()
The Sunburst plot and Sankey diagram are limited in that they do not
show anything about duration of events. The
plotEventDuration() function is an attempt to also provide
something meaningful about the duration of events:
plotEventDuration(
eventDurations = eventDuration,
minCellCount = 5,
treatmentGroups = "both",
eventLines = NULL,
includeOverall = TRUE
)
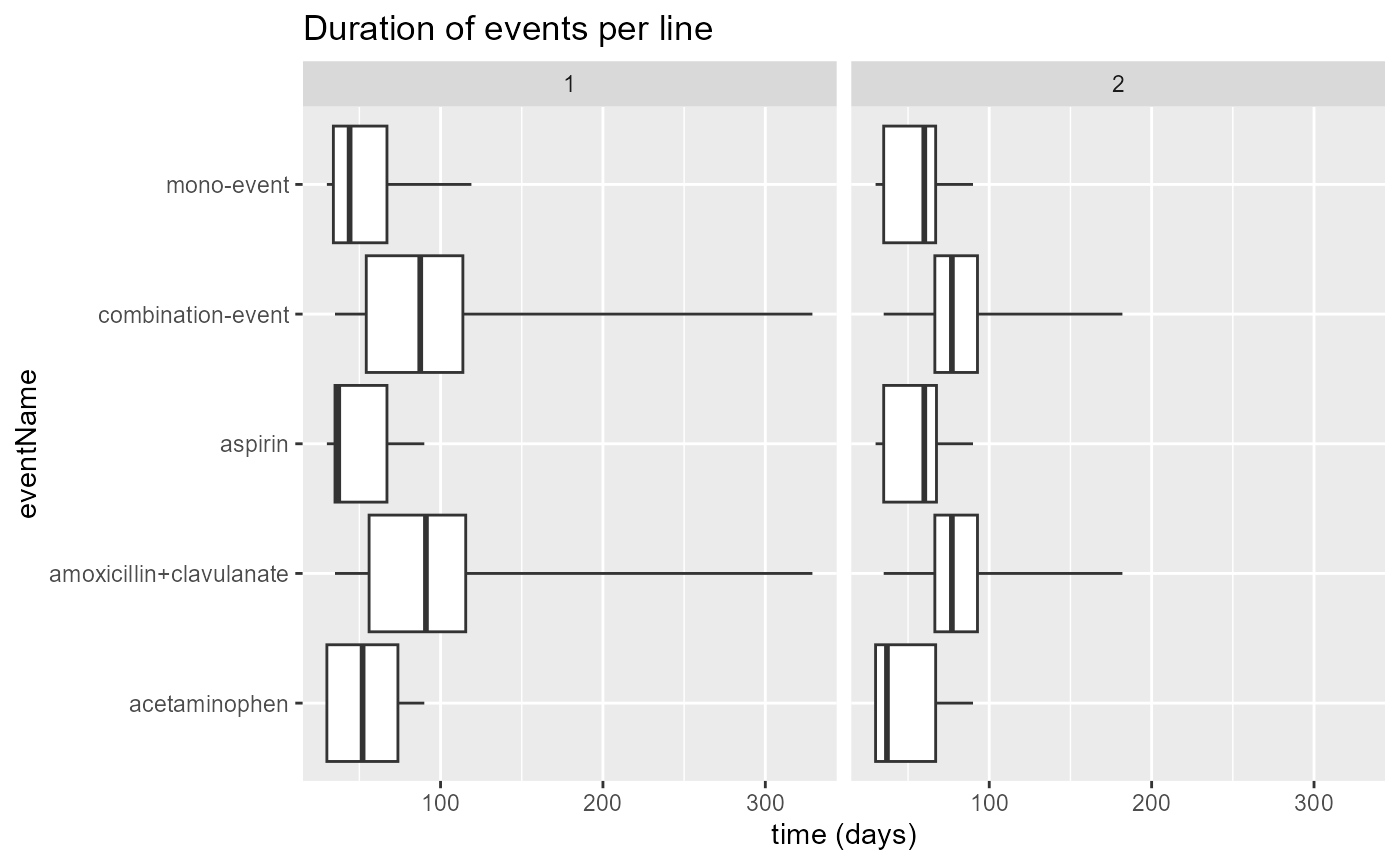
plotEventDuration(
eventDurations = eventDuration,
minCellCount = 5,
treatmentGroups = "both",
eventLines = NULL,
includeOverall = FALSE
)
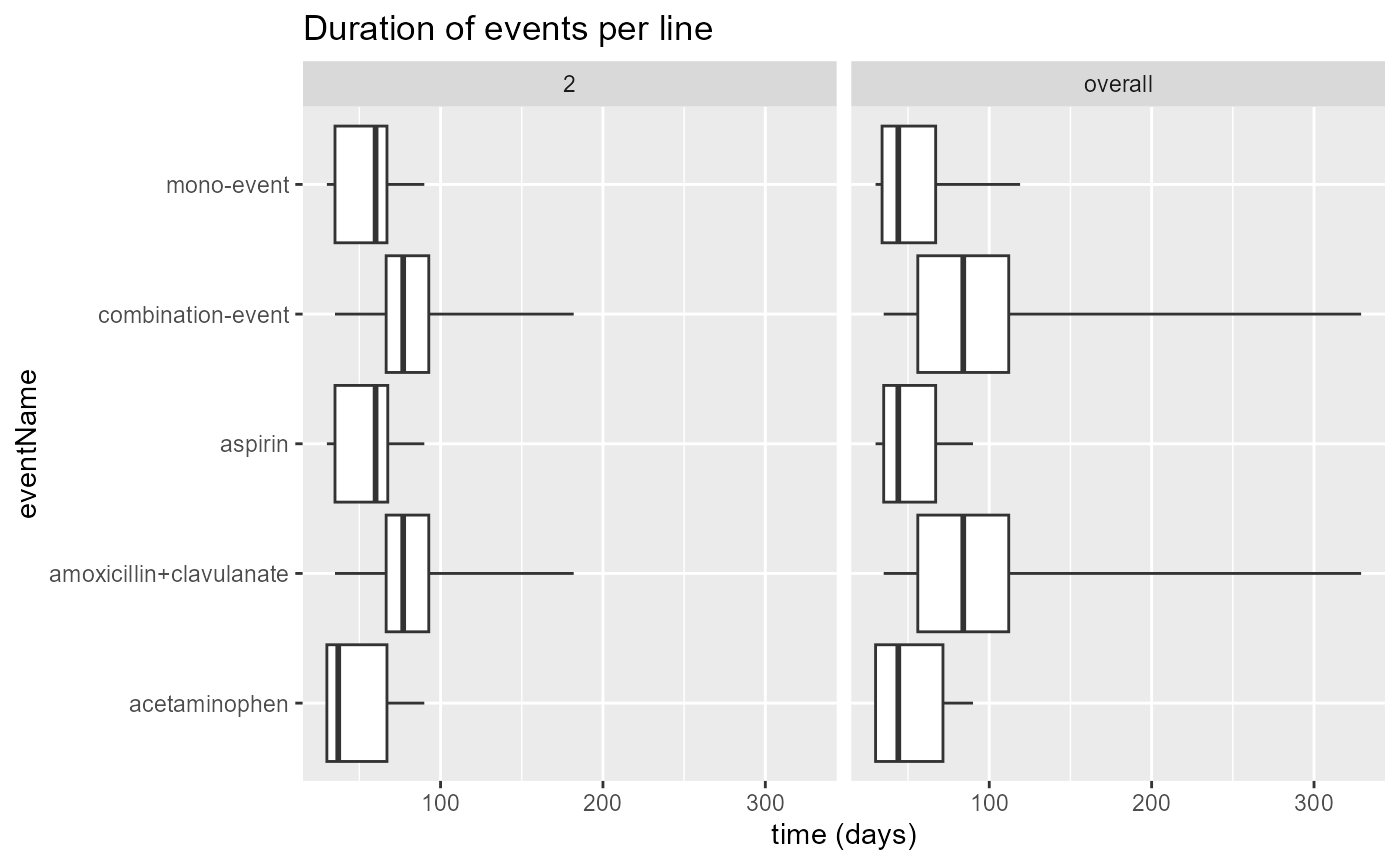
plotEventDuration(
eventDurations = eventDuration,
minCellCount = 5,
treatmentGroups = "both",
eventLines = c(2),
includeOverall = TRUE
)
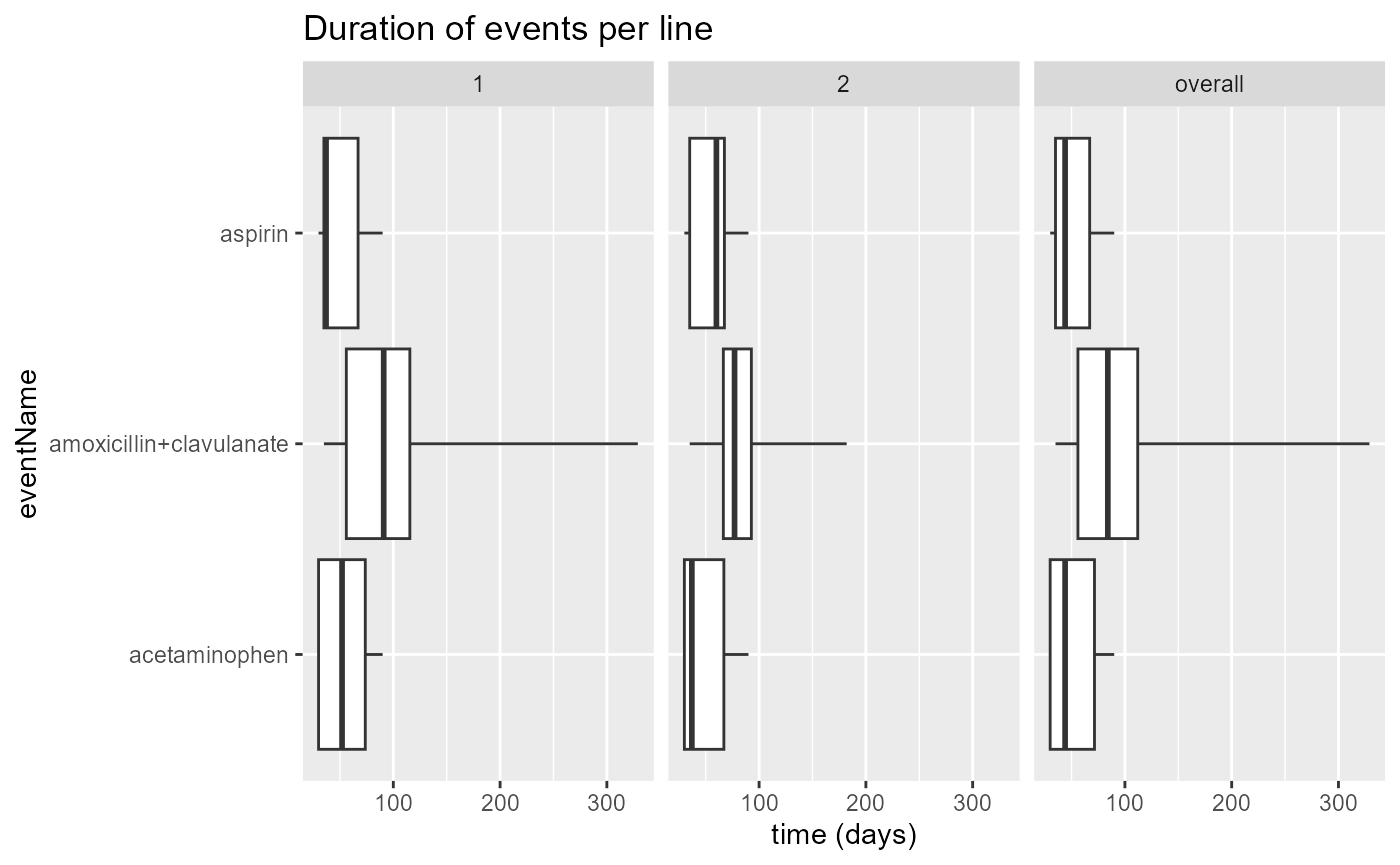
plotEventDuration(
eventDurations = eventDuration,
minCellCount = 5,
treatmentGroups = "individual",
eventLines = NULL,
includeOverall = TRUE
)
plotEventDuration(
eventDurations = eventDuration,
minCellCount = 5,
treatmentGroups = "group",
eventLines = NULL,
includeOverall = TRUE
)
Attrition
Status messages have been updated when running
computePathways(). All messages are now also recorded in an
attrition table. Which keeps track of the number of records and subjects
left in the analysis.
library(TreatmentPatterns)
library(CDMConnector)
library(dplyr)
con <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomia_dir())
cdm <- cdmFromCon(con, cdmSchema = "main", writeSchema = "main")
cohortSet <- readCohortSet(
path = system.file(package = "TreatmentPatterns", "exampleCohorts")
)
cdm <- generateCohortSet(
cdm = cdm,
cohortSet = cohortSet,
name = "cohort_table"
)## ℹ Generating 8 cohorts## ℹ Generating cohort (1/8) - acetaminophen✔ Generating cohort (1/8) - acetaminophen [155ms]
## ℹ Generating cohort (2/8) - amoxicillin✔ Generating cohort (2/8) - amoxicillin [155ms]
## ℹ Generating cohort (3/8) - aspirin✔ Generating cohort (3/8) - aspirin [148ms]
## ℹ Generating cohort (4/8) - clavulanate✔ Generating cohort (4/8) - clavulanate [146ms]
## ℹ Generating cohort (5/8) - death✔ Generating cohort (5/8) - death [105ms]
## ℹ Generating cohort (6/8) - doxylamine✔ Generating cohort (6/8) - doxylamine [150ms]
## ℹ Generating cohort (7/8) - penicillinv✔ Generating cohort (7/8) - penicillinv [154ms]
## ℹ Generating cohort (8/8) - viralsinusitis✔ Generating cohort (8/8) - viralsinusitis [233ms]## Warning: ! 5 casted column in cohort_table (cohort_attrition) as do not match expected
## column type:
## • `number_records` from numeric to integer
## • `number_subjects` from numeric to integer
## • `reason_id` from numeric to integer
## • `excluded_records` from numeric to integer
## • `excluded_subjects` from numeric to integer## Warning: ! 1 column in cohort_table do not match expected column type:
## • `subject_id` is numeric but expected integer
# convert subject_id to subject_id + 1e16 as 64 bit integer
cdm$cohort_table <- cdm$cohort_table %>%
mutate(subject_id = bit64::as.integer64(.data$subject_id + 1e16)) %>%
compute()
cdm$person <- cdm$person %>%
mutate(person_id = bit64::as.integer64(.data$person_id + 1e16)) %>%
compute()
cohorts <- cohortSet %>%
# Remove 'cohort' and 'json' columns
select(-"cohort", -"json") %>%
mutate(type = c("event", "event", "event", "event", "exit", "event", "event", "target")) %>%
rename(
cohortId = "cohort_definition_id",
cohortName = "cohort_name",
) %>%
select("cohortId", "cohortName", "type")
outputEnv <- computePathways(
cohorts = cohorts,
cohortTableName = "cohort_table",
cdm = cdm
)## -- Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## Records: 14041
## Subjects: 2693
## -- Removing records < minEraDuration (0)
## Records: 11386
## Subjects: 2159
## -- Removing events where index date < target index date + indexDateOffset (0)
## Records: 8381
## Subjects: 2159
## -- splitEventCohorts
## Records: 8366
## Subjects: 2144
## -- eraCollapse (30)
## Records: 8366
## Subjects: 2144
## -- Iteration 1: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 558
## Subjects: 512
## -- Iteration 2: minPostCombinationDuration (30), combinatinoWindow (30)
## Records: 554
## Subjects: 512
## -- After Combination
## Records: 554
## Subjects: 512
## -- filterTreatments (First)
## Records: 553
## Subjects: 512
## -- treatment construction done
## Records: 553
## Subjects: 512## Writing attrition to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/attrition.csv
## Writing metadata to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/metadata.csv
## Writing treatmentPathways to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/treatmentPathways.csv
## Censoring 1219 pathways with a frequency <5 to 5.
## Writing summaryStatsTherapyDuration to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/summaryEventDuration.csv
## Writing countsYearPath to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/countsYear.csv
## Writing countsAgePath to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/countsAge.csv
## Writing countsSexPath to C:\Users\MVANKE~1\AppData\Local\Temp\RtmpUjLffZ/tp/countsSex.csv## number_records number_subject reason_id
## 1 14041 2693 1
## 2 11386 2159 2
## 3 8381 2159 3
## 4 8366 2144 4
## 5 8366 2144 5
## 6 558 512 6
## 7 554 512 6
## 8 554 512 7
## 9 553 512 8
## 10 553 512 9
## reason
## 1 Qualifying records for cohort definitions: 1, 2, 3, 4, 5, 6, 7, 8
## 2 Removing records < minEraDuration (0)
## 3 Removing events where index date < target index date + indexDateOffset (0)
## 4 splitEventCohorts
## 5 eraCollapse (30)
## 6 Iteration 1: minPostCombinationDuration (30), combinatinoWindow (30)
## 7 Iteration 2: minPostCombinationDuration (30), combinatinoWindow (30)
## 8 After Combination
## 9 filterTreatments (First)
## 10 treatment construction done
## time
## 1 1732631613
## 2 1732631613
## 3 1732631614
## 4 1732631614
## 5 1732631614
## 6 1732631616
## 7 1732631618
## 8 1732631619
## 9 1732631619
## 10 1732631620The attrition table has the following columns: - number_records: Number of records left in the analysis, at any given stage. - number_subjects: Number of subjects in the analysis, at any given stage. - reason_id: Identification field per stage of the analysis. - reason: Description of why records were removed. - time: Time stamp as in number of seconds since 1970-01-01 (time since epoch).